

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124968.5 - phase: 0
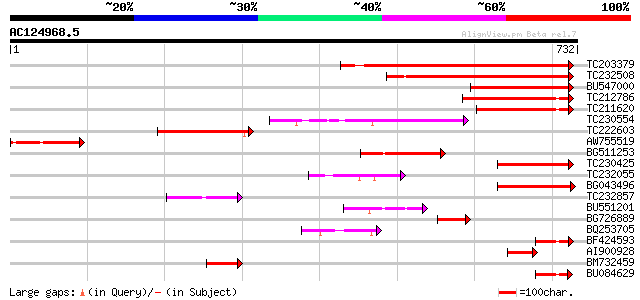
(732 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC203379 similar to GB|BAC55960.1|28071265|AB087408 actin bindin... 346 2e-95
TC232508 similar to GB|BAC55960.1|28071265|AB087408 actin bindin... 290 1e-78
BU547000 similar to GP|29467527|db P0582D05.17 {Oryza sativa (ja... 232 4e-61
TC212786 similar to UP|O49524 (O49524) Pherophorin - like protei... 176 3e-44
TC211620 similar to UP|O49524 (O49524) Pherophorin - like protei... 138 8e-33
TC230554 similar to UP|Q9SX62 (Q9SX62) F11A17.16, partial (24%) 137 1e-32
TC222603 similar to GB|BAC55960.1|28071265|AB087408 actin bindin... 114 1e-25
AW755519 similar to GP|7291109|gb| CG2989-PA {Drosophila melanog... 112 4e-25
BG511253 similar to PIR|G96522|G965 F11A17.16 [imported] - Arabi... 102 6e-22
TC230425 weakly similar to UP|Q93ZL7 (Q93ZL7) At1g48280/F11A17_2... 91 1e-18
TC232055 similar to UP|O49524 (O49524) Pherophorin - like protei... 84 2e-16
BG043496 weakly similar to GP|15983793|gb| At1g48280/F11A17_25 {... 84 3e-16
TC232857 similar to UP|Q96258 (Q96258) AR791, partial (14%) 67 2e-11
BU551201 similar to GP|3261822|emb| PE_PGRS {Mycobacterium tuber... 64 3e-10
BG726889 58 1e-08
BQ253705 weakly similar to GP|27817931|db P0453E05.6 {Oryza sati... 57 3e-08
BF424593 homologue to GP|22135814|gb| AT4g18560/F28J12_220 {Arab... 54 2e-07
AI900928 homologue to GP|28071265|db actin binding protein {Arab... 52 9e-07
BM732459 similar to GP|1669599|dbj| AR791 {Arabidopsis thaliana}... 52 1e-06
BU084629 homologue to GP|22135814|gb| AT4g18560/F28J12_220 {Arab... 49 8e-06
>TC203379 similar to GB|BAC55960.1|28071265|AB087408 actin binding protein
{Arabidopsis thaliana;} , partial (29%)
Length = 1246
Score = 346 bits (887), Expect = 2e-95
Identities = 180/306 (58%), Positives = 222/306 (71%), Gaps = 5/306 (1%)
Frame = +2
Query: 428 QCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPP--PPPPPPPMSFASRGNTA-- 483
+C+ P PPP P A PPPP PPPPPPP SRG
Sbjct: 8 KCSPSNEVPSAPPPPPPPP-----------GAPPPPPPPGGPPPPPPPPGSLSRGGMDGD 154
Query: 484 MVKRAPQVVELYHSLMKRDSRRDSSSGGL-SDAPDVADVRSSMIGEIENRSSHLLAIKAD 542
V RAPQ+VE Y +LMKR++++D+SS L + A + +D RS+MIGEIENRSS LLA+KAD
Sbjct: 155 KVHRAPQLVEFYQTLMKREAKKDTSSSLLVTSASNASDARSNMIGEIENRSSFLLAVKAD 334
Query: 543 IETQGEFVNSLIREVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLKHFDWPEKKADTL 602
+ETQG+FV SL EV A + +I+D+VAFV WLD+EL FLVDERAVLKHFDWPE KAD L
Sbjct: 335 VETQGDFVMSLAAEVRAASFSDINDLVAFVNWLDEELSFLVDERAVLKHFDWPEGKADAL 514
Query: 603 REAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTRDSLMRNC 662
REAAF YQDL KLE+ VS++ DDP LPC+ ALKKM +L EK+E++VY LLRTRD +
Sbjct: 515 REAAFEYQDLMKLENRVSTFVDDPNLPCEAALKKMYSLLEKVEQSVYALLRTRDMAISRY 694
Query: 663 KEFQIPVEWMLDNGIIGKIKLGSVKLAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRF 722
KEF IPV W++D+G++GKIKL SV+LAKKYMKRVA E+ S DK+PA +++VLQGVRF
Sbjct: 695 KEFGIPVNWLMDSGVVGKIKLSSVQLAKKYMKRVASELDELSGPDKEPAREFLVLQGVRF 874
Query: 723 AFRIHQ 728
AFR+HQ
Sbjct: 875 AFRVHQ 892
>TC232508 similar to GB|BAC55960.1|28071265|AB087408 actin binding protein
{Arabidopsis thaliana;} , partial (25%)
Length = 1050
Score = 290 bits (743), Expect = 1e-78
Identities = 143/242 (59%), Positives = 185/242 (76%)
Frame = +2
Query: 487 RAPQVVELYHSLMKRDSRRDSSSGGLSDAPDVADVRSSMIGEIENRSSHLLAIKADIETQ 546
RAP++VE Y +LMK ++++D+S +S D RS+MIGEIENRSS LLA+KAD+ETQ
Sbjct: 2 RAPELVEFYQTLMKLEAKKDTSL--ISSTTYPFDARSNMIGEIENRSSFLLAVKADVETQ 175
Query: 547 GEFVNSLIREVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLKHFDWPEKKADTLREAA 606
G+FV SL EV A + I+D+VAFV WLD+EL FLVDE+AVLKHFDWPE K D +REAA
Sbjct: 176 GDFVISLATEVRAASFSKIEDLVAFVNWLDEELSFLVDEQAVLKHFDWPEGKTDAMREAA 355
Query: 607 FGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTRDSLMRNCKEFQ 666
F Y DL KLE +VS++ DDP+LPC AL+KM +L EK+E+++Y LLRTRD + KEF
Sbjct: 356 FEYLDLMKLEKQVSTFTDDPKLPCQTALQKMYSLLEKVEQSIYALLRTRDMAISRYKEFG 535
Query: 667 IPVEWMLDNGIIGKIKLGSVKLAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRFAFRI 726
IPV W+LD G++GKIKL SV+LA YMKRVA E++ S K+P ++++LQGV FAFR+
Sbjct: 536 IPVTWLLDTGLVGKIKLSSVQLANMYMKRVASELEILSGPKKEPTREFLILQGVHFAFRV 715
Query: 727 HQ 728
HQ
Sbjct: 716 HQ 721
>BU547000 similar to GP|29467527|db P0582D05.17 {Oryza sativa (japonica
cultivar-group)}, partial (19%)
Length = 695
Score = 232 bits (592), Expect = 4e-61
Identities = 116/133 (87%), Positives = 123/133 (92%)
Frame = -2
Query: 596 EKKADTLREAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTR 655
EKKADTLREAAFGYQDLKKLE EV YKDDPRLP DIA KKMVALSEKMERTVY LLRTR
Sbjct: 694 EKKADTLREAAFGYQDLKKLEXEVXXYKDDPRLPGDIAXKKMVALSEKMERTVYNLLRTR 515
Query: 656 DSLMRNCKEFQIPVEWMLDNGIIGKIKLGSVKLAKKYMKRVAIEVQTKSAFDKDPAMDYM 715
DSLMR+ KEF+IP+EWMLDNGIIGKIKL SVKLAKKYMKRVA+E+Q KSA +KDPAMDYM
Sbjct: 514 DSLMRHSKEFKIPIEWMLDNGIIGKIKLSSVKLAKKYMKRVAMELQVKSALEKDPAMDYM 335
Query: 716 VLQGVRFAFRIHQ 728
+LQGVRFAFRIHQ
Sbjct: 334 LLQGVRFAFRIHQ 296
>TC212786 similar to UP|O49524 (O49524) Pherophorin - like protein, partial
(22%)
Length = 430
Score = 176 bits (447), Expect = 3e-44
Identities = 89/144 (61%), Positives = 106/144 (72%)
Frame = +3
Query: 585 ERAVLKHFDWPEKKADTLREAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKM 644
ERAVLKHFDWPE+KAD LREAAFGY DLKKLESE SS++DDPR PC ALKKM L EK+
Sbjct: 3 ERAVLKHFDWPEQKADALREAAFGYCDLKKLESEASSFRDDPRQPCGPALKKMQVLFEKL 182
Query: 645 ERTVYTLLRTRDSLMRNCKEFQIPVEWMLDNGIIGKIKLGSVKLAKKYMKRVAIEVQTKS 704
E V+ + R R+S K F IPV WMLDNG + ++KL SVKLA KYM+RV+ E++T
Sbjct: 183 EHGVFNISRMRESATNRYKVFHIPVHWMLDNGFVSQMKLASVKLAMKYMRRVSAELETGG 362
Query: 705 AFDKDPAMDYMVLQGVRFAFRIHQ 728
P + +V+QGVRFAFR HQ
Sbjct: 363 GC--GPEEEEIVVQGVRFAFRAHQ 428
>TC211620 similar to UP|O49524 (O49524) Pherophorin - like protein, partial
(24%)
Length = 676
Score = 138 bits (348), Expect = 8e-33
Identities = 71/126 (56%), Positives = 90/126 (71%)
Frame = +2
Query: 603 REAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTRDSLMRNC 662
REAAFGY DLKKL SE SS++DDPR C ALKKM L EK+E +Y + R R+S +
Sbjct: 2 REAAFGYCDLKKLVSEASSFRDDPRQLCGPALKKMQTLLEKLEHGIYNISRMRESATKRY 181
Query: 663 KEFQIPVEWMLDNGIIGKIKLGSVKLAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRF 722
K FQIPV+WMLD+G + +IKL SVKLA KYMKRV+ E++T P + +++Q VRF
Sbjct: 182 KVFQIPVDWMLDSGYVSQIKLASVKLAMKYMKRVSAELETGGG---GPEEEELIVQVVRF 352
Query: 723 AFRIHQ 728
AFR+HQ
Sbjct: 353 AFRVHQ 370
>TC230554 similar to UP|Q9SX62 (Q9SX62) F11A17.16, partial (24%)
Length = 1106
Score = 137 bits (346), Expect = 1e-32
Identities = 95/283 (33%), Positives = 138/283 (48%), Gaps = 26/283 (9%)
Frame = +2
Query: 336 SFNLVKKPKKWPITSSDQLSQVECTNNSIIDKNW------------IESISEGSNRRRHS 383
S NL+K + + ++L +V+ N + +N I ++ G+N
Sbjct: 296 SENLIKSLQSEVLALREELDRVKSLNVELESRNTKLTQNLAAAEAKISTVDIGNN----- 460
Query: 384 ISGSNSSEEEVSVLSKRRQSNCFDSFECLKEIEKESVPMPLFVQQCALEKRALRIPNPPP 443
G E S K Q + E ++KE P +F K ++ P P
Sbjct: 461 --GKGPIGEHQSPKFKDIQKLIAEKLE-RSRVKKEGTPEIIFA------KASISAPTPSY 613
Query: 444 RPSCSISSKTKQECSAQVQPPPP--------------PPPPPPPMSFASRGNTAMVKRAP 489
+ S K + +QPPPP PPPPPP+ A +++P
Sbjct: 614 AIPETTSIGRKSPPNTCLQPPPPVTSVGRKSPSNTCLQPPPPPPIPTRPLARLANSQKSP 793
Query: 490 QVVELYHSLMKRDSRRDSSSGGLSDAPDVADVRSSMIGEIENRSSHLLAIKADIETQGEF 549
+VEL+HSL +D + DS P V SS++GEI+NRS+HLLAI+ADIET+GEF
Sbjct: 794 AIVELFHSLKNKDWKIDSKGSVNHQRPVVISAHSSIVGEIQNRSAHLLAIRADIETKGEF 973
Query: 550 VNSLIREVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLKHF 592
+N LIR+V DA + +I++V+ FV WLD +L L DERAVLK F
Sbjct: 974 INDLIRKVVDAAFTDIEEVLKFVDWLDVKLSSLADERAVLKPF 1102
>TC222603 similar to GB|BAC55960.1|28071265|AB087408 actin binding protein
{Arabidopsis thaliana;} , partial (27%)
Length = 881
Score = 114 bits (286), Expect = 1e-25
Identities = 66/130 (50%), Positives = 85/130 (64%), Gaps = 6/130 (4%)
Frame = +2
Query: 192 ERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDIVAK 251
+ + E K + +LE+ VVEL+R NKEL +KR LT +L+ ES+ + N +ES++VAK
Sbjct: 26 DAEVEKKLKAVNDLEVAVVELKRKNKELQHEKRELTVKLNVAESRAAELSNMTESEMVAK 205
Query: 252 FKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKNT------CSTL 305
K E S LR NEDL KQVEGLQ +R +EVEEL YLRWVN+CLR EL+N S
Sbjct: 206 AKEEVSNLRHANEDLLKQVEGLQMNRFSEVEELVYLRWVNACLRYELRNNQTPQGKVSAR 385
Query: 306 DSDKLSSPQS 315
D K SP+S
Sbjct: 386 DLSKSLSPKS 415
>AW755519 similar to GP|7291109|gb| CG2989-PA {Drosophila melanogaster},
partial (0%)
Length = 433
Score = 112 bits (281), Expect = 4e-25
Identities = 66/100 (66%), Positives = 71/100 (71%), Gaps = 4/100 (4%)
Frame = +2
Query: 1 MKEEIHNNPSENKTKVSKFSDQNQPPKLQTTKTTNPNNNNHSKPRLWGAHIVKGFSADKK 60
MKEE NPSEN++K SKFSDQNQ PK Q TK N NNNN K RLWGAHIVKGFSADKK
Sbjct: 149 MKEE---NPSENRSKASKFSDQNQAPKPQNTKGANLNNNN--KTRLWGAHIVKGFSADKK 313
Query: 61 TKQLPTKK--QQNTTTITTSDVVTNQKNVN-PFVP-PHSR 96
TK +KK TTT + +VVTNQ N N PFVP HSR
Sbjct: 314 TKSFASKKLTSSTTTTAISENVVTNQNNSNKPFVPSSHSR 433
>BG511253 similar to PIR|G96522|G965 F11A17.16 [imported] - Arabidopsis
thaliana, partial (6%)
Length = 433
Score = 102 bits (254), Expect = 6e-22
Identities = 48/109 (44%), Positives = 73/109 (66%)
Frame = +2
Query: 454 KQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAPQVVELYHSLMKRDSRRDSSSGGLS 513
K C++ + PPPPP PP P ++ N +RAP V+L+H+L ++ + ++ G
Sbjct: 107 KPTCNSCLPPPPPPMPPSIPSRPIAKANN--TQRAPAFVKLFHTLKNQEGMKSTTGSGKQ 280
Query: 514 DAPDVADVRSSMIGEIENRSSHLLAIKADIETQGEFVNSLIREVNDAVY 562
P +V SS++GEI+NRS+HLLAI+ADIET+GEF+N LI++V +A Y
Sbjct: 281 QRPVAVNVHSSIVGEIQNRSAHLLAIRADIETKGEFINDLIKKVVEAAY 427
>TC230425 weakly similar to UP|Q93ZL7 (Q93ZL7) At1g48280/F11A17_25, partial
(61%)
Length = 784
Score = 91.3 bits (225), Expect = 1e-18
Identities = 42/99 (42%), Positives = 66/99 (66%)
Frame = +1
Query: 630 CDIALKKMVALSEKMERTVYTLLRTRDSLMRNCKEFQIPVEWMLDNGIIGKIKLGSVKLA 689
C ALKKM +L +K ER++ L++ R S+ + + + IP WMLD+GI+ +IK S+ L
Sbjct: 22 CGAALKKMASLLDKSERSIQRLIKLRSSVTHSYQMYNIPTAWMLDSGIMSEIKQASMTLV 201
Query: 690 KKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRFAFRIHQ 728
K YMKRV +E+++ D++ D ++LQG+ FA+R HQ
Sbjct: 202 KTYMKRVTMELESIRNSDRESIQDSLLLQGMHFAYRAHQ 318
>TC232055 similar to UP|O49524 (O49524) Pherophorin - like protein, partial
(7%)
Length = 448
Score = 84.3 bits (207), Expect = 2e-16
Identities = 59/147 (40%), Positives = 77/147 (52%), Gaps = 22/147 (14%)
Frame = +2
Query: 386 GSNSSEEEVSVLSKR-RQSNCFDSFECLKEIEKESVPMPLFVQQCALEKRALRIPNPPPR 444
GS + ++EVS +S+R R S C +E V V ++ RA R+PNPPP+
Sbjct: 17 GSGNLKKEVSEISERPRHSRCNS---------EELVDCSDSVLFGSVGSRAPRVPNPPPK 169
Query: 445 PSCSIS-----SKTKQECSAQVQPPPPPPP--------------PPPPMSFASRGNTAMV 485
P+ S S S + E ++ PPPPPPP PPPP + A R A V
Sbjct: 170 PTSSSSPSSPSSVSSGETERRIPPPPPPPPRKPASNAAAAPPPPPPPPPAKAVRVAPAKV 349
Query: 486 KRAPQVVELYHSLMKR--DSRRDSSSG 510
+R P+V E YHSLM+R DSRRDS SG
Sbjct: 350 RRVPEVEEFYHSLMRRDSDSRRDSGSG 430
>BG043496 weakly similar to GP|15983793|gb| At1g48280/F11A17_25 {Arabidopsis
thaliana}, partial (43%)
Length = 423
Score = 83.6 bits (205), Expect = 3e-16
Identities = 38/101 (37%), Positives = 65/101 (63%)
Frame = +1
Query: 630 CDIALKKMVALSEKMERTVYTLLRTRDSLMRNCKEFQIPVEWMLDNGIIGKIKLGSVKLA 689
C + L + + ++ ER++ L++ R S+ + + + IP WMLD+GI+ +IK S+ L
Sbjct: 13 CMLTLSLISSHCDRSERSIQRLIKLRSSVTHSYQMYNIPTAWMLDSGIMSEIKQASMTLV 192
Query: 690 KKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRFAFRIHQVF 730
K YMKRV +E+++ D++ D ++LQG+ FA+R HQVF
Sbjct: 193 KTYMKRVTMELESIRNSDRESIQDSLLLQGMHFAYRAHQVF 315
>TC232857 similar to UP|Q96258 (Q96258) AR791, partial (14%)
Length = 892
Score = 67.4 bits (163), Expect = 2e-11
Identities = 37/98 (37%), Positives = 57/98 (57%)
Frame = +1
Query: 203 QNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLT 262
++LE E ELR+ N L ++ +L RL S + + E+ V K E+ L+
Sbjct: 442 KDLESEAEELRKSNLRLQIENSDLARRLDSTQILANAFLEDPEAGAV---KQESECLKQE 612
Query: 263 NEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKN 300
N L K++E Q+ R +++EEL YLRW+N+CLR EL+N
Sbjct: 613 NVRLMKEIEQFQSDRCSDLEELVYLRWINACLRYELRN 726
>BU551201 similar to GP|3261822|emb| PE_PGRS {Mycobacterium tuberculosis
H37Rv}, partial (7%)
Length = 689
Score = 63.5 bits (153), Expect = 3e-10
Identities = 40/114 (35%), Positives = 62/114 (54%), Gaps = 5/114 (4%)
Frame = +3
Query: 431 LEKRALRIPNPPPRPSCSISSKTKQECSAQVQP-----PPPPPPPPPPMSFASRGNTAMV 485
++ R++++P P P + + S K E +VQP PPPPP PP S + V
Sbjct: 129 VKDRSVKVPPPAPSSNPLLPSH-KTEKGMKVQPLALPRTAPPPPPTPPKSLVGLKS---V 296
Query: 486 KRAPQVVELYHSLMKRDSRRDSSSGGLSDAPDVADVRSSMIGEIENRSSHLLAI 539
+R P+V+ELY SL ++D+ D+ ++ A +MI EIENRS+ L A+
Sbjct: 297 RRVPEVIELYRSLTRKDANNDNKIS--TNGTPAAAFTRNMIEEIENRSTFLSAV 452
>BG726889
Length = 396
Score = 58.2 bits (139), Expect = 1e-08
Identities = 28/44 (63%), Positives = 33/44 (74%), Gaps = 1/44 (2%)
Frame = -1
Query: 553 LIREVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLKHF-DWP 595
L++EV A Y +I +V AFVKWLD EL LVDER+VLKHF WP
Sbjct: 132 LLKEVESATYADISEVEAFVKWLDGELSSLVDERSVLKHFPHWP 1
>BQ253705 weakly similar to GP|27817931|db P0453E05.6 {Oryza sativa (japonica
cultivar-group)}, partial (2%)
Length = 403
Score = 57.0 bits (136), Expect = 3e-08
Identities = 39/115 (33%), Positives = 55/115 (46%), Gaps = 12/115 (10%)
Frame = +2
Query: 377 SNRRRHSISGSNSSEEEVSVLSKR---RQSNCFDSFECLKEIEKESVPMPLFVQQCALEK 433
S ++ S GS + + EVS +S+R + N + +C + SV
Sbjct: 59 SLKKTASDLGSGNLKREVSEISERPLHSRCNSEELVDCSDSVLSGSV-----------RS 205
Query: 434 RALRIPNPPPRPSCSI-SSKTKQECSAQVQPPP--------PPPPPPPPMSFASR 479
RA R+PNPPP+PS S SS + E ++ PPP PPPPPPPP S+
Sbjct: 206 RAPRVPNPPPKPSTSSPSSGSSGETERRIPPPPPPPPMKALPPPPPPPPKKLVSK 370
>BF424593 homologue to GP|22135814|gb| AT4g18560/F28J12_220 {Arabidopsis
thaliana}, partial (11%)
Length = 411
Score = 53.9 bits (128), Expect = 2e-07
Identities = 27/49 (55%), Positives = 36/49 (73%)
Frame = +3
Query: 680 KIKLGSVKLAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRFAFRIHQ 728
+IKL SVKLA KYMKRV+ E++T P + +++QGVRFAFR+HQ
Sbjct: 111 QIKLASVKLAMKYMKRVSAELETGGG---GPEEEELIVQGVRFAFRVHQ 248
>AI900928 homologue to GP|28071265|db actin binding protein {Arabidopsis
thaliana}, partial (3%)
Length = 425
Score = 52.0 bits (123), Expect = 9e-07
Identities = 20/39 (51%), Positives = 31/39 (79%)
Frame = -2
Query: 643 KMERTVYTLLRTRDSLMRNCKEFQIPVEWMLDNGIIGKI 681
++E++VY LLRTRD + KEF IPV W++D+G++GK+
Sbjct: 265 RVEQSVYALLRTRDMAISRYKEFGIPVNWLMDSGVVGKV 149
>BM732459 similar to GP|1669599|dbj| AR791 {Arabidopsis thaliana}, partial
(12%)
Length = 435
Score = 51.6 bits (122), Expect = 1e-06
Identities = 23/46 (50%), Positives = 32/46 (69%)
Frame = +3
Query: 255 EASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKN 300
E LR NE L+K++E L R ++EEL YLRW+N+CLR EL++
Sbjct: 3 EGERLRRENEGLTKELEQLHADRCLDLEELVYLRWINACLRHELRS 140
>BU084629 homologue to GP|22135814|gb| AT4g18560/F28J12_220 {Arabidopsis
thaliana}, partial (6%)
Length = 420
Score = 48.9 bits (115), Expect = 8e-06
Identities = 25/47 (53%), Positives = 34/47 (72%)
Frame = +2
Query: 680 KIKLGSVKLAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRFAFRI 726
+IKL SVKLA KYMKRV+ E++T P + +++QGVRFAFR+
Sbjct: 287 QIKLASVKLAMKYMKRVSAELETGGG---GPEEEELIVQGVRFAFRV 418
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.128 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,210,009
Number of Sequences: 63676
Number of extensions: 635176
Number of successful extensions: 26427
Number of sequences better than 10.0: 905
Number of HSP's better than 10.0 without gapping: 7152
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 14393
length of query: 732
length of database: 12,639,632
effective HSP length: 104
effective length of query: 628
effective length of database: 6,017,328
effective search space: 3778881984
effective search space used: 3778881984
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC124968.5


