

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124967.4 - phase: 0 /pseudo
(335 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
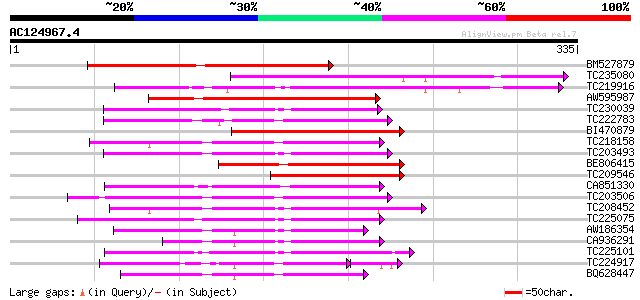
Score E
Sequences producing significant alignments: (bits) Value
BM527879 weakly similar to GP|9758483|dbj pectin methylesterase-... 173 1e-43
TC235080 132 2e-31
TC219916 similar to GB|BAB01985.1|9294028|AB018121 pectin methyl... 132 3e-31
AW595987 114 5e-26
TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase i... 109 2e-24
TC222783 weakly similar to UP|Q9SMV9 (Q9SMV9) Pectin methylester... 107 6e-24
BI470879 similar to GP|20856815|gb AT5g47500/MNJ7_9 {Arabidopsis... 106 1e-23
TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pe... 103 1e-22
TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like prote... 98 6e-21
BE806415 similar to GP|9294028|dbj pectin methylesterase-like pr... 97 1e-20
TC209546 similar to UP|Q8LPF3 (Q8LPF3) AT5g47500/MNJ7_9, partial... 96 2e-20
CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.... 95 5e-20
TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (... 94 1e-19
TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (... 89 3e-18
TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 88 5e-18
AW186354 85 5e-17
CA936291 82 3e-16
TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 82 3e-16
TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4... 73 7e-16
BQ628447 similar to GP|10178067|db pectin methylesterase-like pr... 80 2e-15
>BM527879 weakly similar to GP|9758483|dbj pectin methylesterase-like protein
{Arabidopsis thaliana}, partial (36%)
Length = 422
Score = 173 bits (438), Expect = 1e-43
Identities = 81/145 (55%), Positives = 110/145 (75%)
Frame = +1
Query: 47 AETTVRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTF 106
AE +VV+V +DG+G+F T+TDA+KSIPSGN +RV+++IG G Y EKI ++++K FVT
Sbjct: 1 AEEGAKVVKVMQDGSGEFKTITDAIKSIPSGNTKRVIIYIGAGNYNEKIKIEKTKPFVTL 180
Query: 107 YGERNGKDNDMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGG 166
YG + MP +T+ TA +YGT+DSAT+ V++DYFVA N+ N++P PD + GG
Sbjct: 181 YGVP-----EKMPNLTFGGTAQQYGTVDSATLIVESDYFVAANIMISNTAPRPDPKTPGG 345
Query: 167 QALAMRISGDKAAFYNCKFIGFQDT 191
QA+A+RISGDKAAFYNCK GFQDT
Sbjct: 346 QAVALRISGDKAAFYNCKMYGFQDT 420
>TC235080
Length = 807
Score = 132 bits (333), Expect = 2e-31
Identities = 78/221 (35%), Positives = 115/221 (51%), Gaps = 21/221 (9%)
Frame = +3
Query: 131 GTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALAMRISGDKAAFYNCKFIGFQD 190
GT SAT AV++ YF+A N+ F N++P+P +VG Q +A+RIS D A F CKF+G QD
Sbjct: 51 GTYGSATFAVNSPYFIAKNITFKNTAPIPAPGAVGKQGVALRISADTAVFLGCKFLGAQD 230
Query: 191 TLCDDYGKHFFKDCFIQGTYDFIFGNGKSIYLRTTIESVAK-----------------GF 233
TL D G+H++KDC+I+G+ DFIFGN S++ + ++A+ GF
Sbjct: 231 TLYDHIGRHYYKDCYIEGSVDFIFGNALSLFEGCHVHAIAQLTGALTAQGRNSLLEDTGF 410
Query: 234 ECHHSTR*GKN---VGRHWIHFCSLQHNWKWSQE-HLPWSWLEEEP*GCICLYLYGLCRQ 289
H G +GR W F + + + +P W YG
Sbjct: 411 SFVHCKVTGSGALYLGRAWGPFSRVVFAYTYMDNIIIPKGWYNWGDPNREMTVFYGQ--- 581
Query: 290 FQRMVPPWVQ*AVRLNYKRILSDEEAKHFLSMAYIHGEQWV 330
+ P A R+++ R LSDEEAK F+S++YI G +W+
Sbjct: 582 -YKCTGPGASYAGRVSWSRELSDEEAKPFISLSYIDGSEWI 701
>TC219916 similar to GB|BAB01985.1|9294028|AB018121 pectin
methylesterase-like protein {Arabidopsis thaliana;} ,
partial (94%)
Length = 1163
Score = 132 bits (331), Expect = 3e-31
Identities = 93/301 (30%), Positives = 148/301 (48%), Gaps = 36/301 (11%)
Frame = +2
Query: 63 DFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYGERNGKDNDMMPIIT 122
DF TV +A+ ++P GN RR V+ + G YR+ + V ++K F+T + +D ++T
Sbjct: 5 DFQTVQEAIDAVPLGNIRRTVIRVSPGIYRQPVYVPKTKNFITL-AALSPEDT----VLT 169
Query: 123 YDATA-----------LRYGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALAM 171
++ TA + GT + V+ + F+A N+ F NS+P E S GQA+A+
Sbjct: 170 WNNTATGIDHHQPARVIGTGTFGCGSTIVEGEDFIAENITFENSAP---EGS--GQAVAI 334
Query: 172 RISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIYLRTTIESVAK 231
R++ D+ AFYNC+F+G+QDTL YGK + KDC+I+G+ DFIFGN ++ I +
Sbjct: 335 RVTADRCAFYNCRFLGWQDTLYLHYGKQYLKDCYIEGSVDFIFGNSTALLEHCHIHCKSA 514
Query: 232 GFECHHSTR*GKN---------------------VGRHWIHFCSLQHNWKWSQE---HLP 267
GF S + + +GR W F + + + + H+
Sbjct: 515 GFITAQSRKSSQETTGYVFLRCVITGNGGNSYAYLGRPWGPFGRVVFAYTYMDQCIRHVG 694
Query: 268 W-SWLEEEP*GCICLYLYGLCRQFQRMVPPWVQ*AVRLNYKRILSDEEAKHFLSMAYIHG 326
W +W + E C Y Y R P + R+ + R L DEEA+ FL+ +I
Sbjct: 695 WDNWGKMENERSACFYEY-------RCFGPGCCPSKRVTWCRELLDEEAEQFLTHPFIDP 853
Query: 327 E 327
E
Sbjct: 854 E 856
>AW595987
Length = 410
Score = 114 bits (286), Expect = 5e-26
Identities = 62/138 (44%), Positives = 86/138 (61%), Gaps = 1/138 (0%)
Frame = +3
Query: 83 VVWIGMGEYREKITVDRSKRFVTFYGERNGKDNDMMPIITYDATALRYG-TLDSATVAVD 141
++ I G YREK+ V +K + G+ + I ++ TA G T S + AV
Sbjct: 12 LIIIDSGTYREKVVVQANKTNLIVQGQ-----GYLNTTIEWNDTANSTGGTSYSYSFAVF 176
Query: 142 ADYFVAVNVAFVNSSPMPDENSVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFF 201
A F A N++F N++P P VG QA+A+R++GD+AAFY C F G QDTL DD G+H+F
Sbjct: 177 ASKFTAYNISFKNTAPPPSPGVVGAQAVALRVTGDQAAFYGCGFYGAQDTLNDDGGRHYF 356
Query: 202 KDCFIQGTYDFIFGNGKS 219
K+CFIQG+ DFIFGN +S
Sbjct: 357 KECFIQGSIDFIFGNARS 410
>TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (70%)
Length = 913
Score = 109 bits (272), Expect = 2e-24
Identities = 65/165 (39%), Positives = 91/165 (54%)
Frame = +1
Query: 56 VRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYGERNGKDN 115
V KDGTG+FTT+ +AV P+ + R V+ I G Y E + V R K + F G+ GK
Sbjct: 235 VAKDGTGNFTTIAEAVAVAPNSSATRFVIHIKAGAYFENVEVIRKKTNLMFVGDGIGKTV 414
Query: 116 DMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALAMRISG 175
D + T SATVAV D F+A + F NS+ P ++ QA+A+R
Sbjct: 415 VKASRNVVDG----WTTFQSATVAVVGDGFIAKGITFENSAG-PSKH----QAVALRSGS 567
Query: 176 DKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSI 220
D +AFY C F+ +QDTL + F++DC + GT DFIFGN ++
Sbjct: 568 DFSAFYKCSFVAYQDTLYVHSLRQFYRDCDVYGTVDFIFGNAATV 702
>TC222783 weakly similar to UP|Q9SMV9 (Q9SMV9) Pectin methylesterase
precursor , partial (29%)
Length = 630
Score = 107 bits (268), Expect = 6e-24
Identities = 63/176 (35%), Positives = 100/176 (56%), Gaps = 5/176 (2%)
Frame = +3
Query: 56 VRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYGERNGKDN 115
V KDG+G FTTV DA+ S P ++ R ++++ G Y E ITVD+ K + YG D
Sbjct: 93 VAKDGSGQFTTVLDAINSYPKKHQGRYIIYVKAGIYDEYITVDKKKPNLLIYG-----DG 257
Query: 116 DMMPIIT-----YDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALA 170
IIT ++ T T+ +AT + A+ F+A ++AF N++ + G QA+A
Sbjct: 258 PTKTIITGRKNFHEGTK----TMRTATFSTVAEDFMAKSIAFENTA-----GAEGHQAVA 410
Query: 171 MRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIYLRTTI 226
+R+ GD++ F++C G+QDTL + F+++C I GT DFIFG ++ + I
Sbjct: 411 LRVQGDRSVFFDCAMRGYQDTLYAHAHRQFYRNCEISGTIDFIFGYSTTLIQNSKI 578
>BI470879 similar to GP|20856815|gb AT5g47500/MNJ7_9 {Arabidopsis thaliana},
partial (39%)
Length = 430
Score = 106 bits (265), Expect = 1e-23
Identities = 51/102 (50%), Positives = 69/102 (67%)
Frame = +2
Query: 132 TLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALAMRISGDKAAFYNCKFIGFQDT 191
T +A+V V A+YF A N++F N++P P G QA A RISG + F C F G QDT
Sbjct: 59 TYRTASVTVFANYFSARNISFKNTAPAPMPGMEGWQAAAFRISGFFSYFSGCGFYGAQDT 238
Query: 192 LCDDYGKHFFKDCFIQGTYDFIFGNGKSIYLRTTIESVAKGF 233
LCDD G+H+FK+C+I+G+ DFIFGNG+S+Y + S+A F
Sbjct: 239 LCDDAGRHYFKECYIEGSIDFIFGNGRSMYKDCRLHSIATRF 364
>TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pectin
methylesterase-like protein, partial (59%)
Length = 1291
Score = 103 bits (257), Expect = 1e-22
Identities = 62/179 (34%), Positives = 101/179 (55%), Gaps = 5/179 (2%)
Frame = +3
Query: 48 ETTVR-VVRVRKDGTGDFTTVTDAVKSIPSGNKRR---VVVWIGMGEYREKITVDRSKRF 103
E VR +V V +DG+G+FTT+ DA+ + P+ + ++++ G Y E +++D+ K +
Sbjct: 132 EVVVRDIVTVSQDGSGNFTTINDAIAAAPNKSVSTDGYFLIYVTAGVYEENVSIDKKKTY 311
Query: 104 VTFYGERNGKDNDMMPIITYDATALR-YGTLDSATVAVDADYFVAVNVAFVNSSPMPDEN 162
+ G+ K IIT + + + + T SAT+AV FV VN+ N++
Sbjct: 312 LMMVGDGINKT-----IITGNRSVVDGWTTFSSATLAVVGQGFVGVNMTIRNTA-----G 461
Query: 163 SVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIY 221
+V QA+A+R D + FY+C F G+QDTL + F+ +C I GT DFIFGN K ++
Sbjct: 462 AVKHQAVALRSGADLSTFYSCSFEGYQDTLYVHSLRQFYSECDIYGTVDFIFGNAKVVF 638
>TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like protein
(At3g49220), partial (59%)
Length = 1298
Score = 97.8 bits (242), Expect = 6e-21
Identities = 57/173 (32%), Positives = 90/173 (51%), Gaps = 2/173 (1%)
Frame = +1
Query: 56 VRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYRE-KITVDRSKRFVTFYGERNGKD 114
V KDG G T+ +A+K +P + RR++++I G Y E + + R K V F G+ GK
Sbjct: 214 VSKDGNGTVKTIAEAIKKVPEYSSRRIIIYIRAGRYEEDNLKLGRKKTNVMFIGDGKGKT 393
Query: 115 NDMMPIITYDATALR-YGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALAMRI 173
+IT + T +A+ A F+A ++ F N + P + QA+A+R+
Sbjct: 394 -----VITGGRNYYQNLTTFHTASFAASGSGFIAKDMTFENYAG-PGRH----QAVALRV 543
Query: 174 SGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIYLRTTI 226
D A Y C IG+QDT+ + F+++C I GT DFIFGN ++ T+
Sbjct: 544 GADHAVVYRCNIIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCTL 702
>BE806415 similar to GP|9294028|dbj pectin methylesterase-like protein
{Arabidopsis thaliana}, partial (41%)
Length = 399
Score = 96.7 bits (239), Expect = 1e-20
Identities = 47/110 (42%), Positives = 67/110 (60%)
Frame = +1
Query: 124 DATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALAMRISGDKAAFYNC 183
DA + GT T+ V+ F+A N+ F NSSP GQA+A+R++ D+ AFYNC
Sbjct: 22 DARVIGTGTFGCGTIIVEGGDFIAENITFENSSPQG-----AGQAVAVRVTVDRCAFYNC 186
Query: 184 KFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIYLRTTIESVAKGF 233
+F+G+QDTL YG + KDC+I+G+ DFIFGN ++ I + GF
Sbjct: 187 RFLGWQDTLYLHYGIQYLKDCYIEGSVDFIFGNSTALLEHCHIHCKSAGF 336
>TC209546 similar to UP|Q8LPF3 (Q8LPF3) AT5g47500/MNJ7_9, partial (55%)
Length = 680
Score = 95.9 bits (237), Expect = 2e-20
Identities = 43/79 (54%), Positives = 56/79 (70%)
Frame = +2
Query: 155 SSPMPDENSVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIF 214
++P P G QA+A RISGDKA F C F G QDTLCDD G+H+FK+C+I+G+ DFIF
Sbjct: 2 TAPAPMPGMQGWQAVAFRISGDKAYFSGCGFYGAQDTLCDDAGRHYFKECYIEGSIDFIF 181
Query: 215 GNGKSIYLRTTIESVAKGF 233
GNG+S+Y + S+A F
Sbjct: 182 GNGRSMYKDCELHSIATRF 238
>CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.150 -
Arabidopsis thaliana, partial (37%)
Length = 700
Score = 94.7 bits (234), Expect = 5e-20
Identities = 58/166 (34%), Positives = 88/166 (52%), Gaps = 1/166 (0%)
Frame = +2
Query: 57 RKDGTGDFTTVTDAVKSIPSGN-KRRVVVWIGMGEYREKITVDRSKRFVTFYGERNGKDN 115
RK G+G+F TV DA+ + K R V+ + G YRE I V + G+ G N
Sbjct: 5 RKXGSGNFKTVQDALNAAAKRKEKTRFVIHVKKGVYRENIEVALHNDNIMLVGD--GLRN 178
Query: 116 DMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALAMRISG 175
+ I + + Y T SAT +D +F+A ++ F NS+ + GQA+A+R +
Sbjct: 179 TI--ITSARSVQDGYTTYSSATAGIDGLHFIARDITFQNSAGVHK-----GQAVALRSAS 337
Query: 176 DKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIY 221
D + FY C +G+QDTL + F++ C+I GT DFIFGN ++
Sbjct: 338 DLSVFYRCGIMGYQDTLMAHAQRQFYRQCYIYGTVDFIFGNAAVVF 475
>TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (57%)
Length = 1320
Score = 93.6 bits (231), Expect = 1e-19
Identities = 60/194 (30%), Positives = 93/194 (47%), Gaps = 2/194 (1%)
Frame = +2
Query: 35 REITLLEEHLTAAETTVRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREK 94
R+ LL L+A + + V K G G T+ +A+K P + RR ++++ G Y E
Sbjct: 32 RDRRLLSLPLSAIQADITV---SKTGNGTVKTIAEAIKKAPEHSTRRFIIYVRAGRYEEN 202
Query: 95 -ITVDRSKRFVTFYGERNGKDNDMMPIITYDATALR-YGTLDSATVAVDADYFVAVNVAF 152
+ V R K V F G+ GK +IT + T SA+ A F+A ++ F
Sbjct: 203 NLKVGRKKTNVMFIGDGKGKT-----VITGKKNVIDGMTTFHSASFAASGAGFIARDITF 367
Query: 153 VNSSPMPDENSVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDF 212
N + QA+A+R+ D A Y C +G+QD+ + FF++C I GT DF
Sbjct: 368 ENYA-----GPAKHQAVALRVGADHAVVYRCNIVGYQDSCYVHSNRQFFRECNIYGTVDF 532
Query: 213 IFGNGKSIYLRTTI 226
IFGN ++ + I
Sbjct: 533 IFGNAAVVFQKCNI 574
>TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (56%)
Length = 1273
Score = 89.0 bits (219), Expect = 3e-18
Identities = 60/197 (30%), Positives = 102/197 (51%), Gaps = 10/197 (5%)
Frame = +1
Query: 60 GTGDFTTVTDAVKSIPSGNKRR---VVVWIGMGEYREKITVDRSKRFVTFYGERNGKDND 116
G ++T++ DA+ + P+ K +V++ G Y E + + + K+ + G+ K
Sbjct: 310 GIDNYTSIGDAIAAAPNNTKPEDGYFLVYVREGLYEEYVVIPKEKKNILLVGDGINKT-- 483
Query: 117 MMPIITYDATALR-YGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALAMRISG 175
IIT + + + + T +S+T AV + F+AV+V F N++ P+++ QA+A+R +
Sbjct: 484 ---IITGNHSVIDGWTTFNSSTFAVSGERFIAVDVTFRNTAG-PEKH----QAVAVRNNA 639
Query: 176 DKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGN------GKSIYLRTTIESV 229
D + FY C F G+QDTL + F+++C I GT DFIFGN G IY R + +
Sbjct: 640 DLSTFYRCSFEGYQDTLYVHSLRQFYRECEIYGTVDFIFGNAAVVFQGCKIYARKPLPNQ 819
Query: 230 AKGFECHHSTR*GKNVG 246
T +N G
Sbjct: 820 KNAVTAQGRTDPNQNTG 870
Score = 28.1 bits (61), Expect = 5.8
Identities = 13/31 (41%), Positives = 18/31 (57%)
Frame = +3
Query: 227 ESVAKGFECHHSTR*GKNVGRHWIHFCSLQH 257
+++AK EC HSTR + +HW LQH
Sbjct: 801 KTIAKPEECCHSTRKN*SKSKHWHFDPKLQH 893
>TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (87%)
Length = 1744
Score = 88.2 bits (217), Expect = 5e-18
Identities = 54/181 (29%), Positives = 92/181 (49%)
Frame = +1
Query: 41 EEHLTAAETTVRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRS 100
E L A V V DG+G++ + DAV + P + +R V+ + G Y E + + +
Sbjct: 580 ERKLLQAIAVTPDVTVALDGSGNYAKIMDAVLAAPDYSMKRFVILVKKGVYVENVEIKKK 759
Query: 101 KRFVTFYGERNGKDNDMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMPD 160
K + G+ D I + + T SAT AV F+A +++F N++ P+
Sbjct: 760 KWNIMILGQ----GMDATVISGNRSVVDGWTTFRSATFAVSGRGFIARDISFQNTAG-PE 924
Query: 161 ENSVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSI 220
++ QA+A+R D + F+ C G+QD+L + FF+DC I GT D+IFG+ ++
Sbjct: 925 KH----QAVALRSDSDLSVFFRCGIFGYQDSLYTHTMRQFFRDCTISGTVDYIFGDATAV 1092
Query: 221 Y 221
+
Sbjct: 1093F 1095
>AW186354
Length = 432
Score = 84.7 bits (208), Expect = 5e-17
Identities = 55/153 (35%), Positives = 83/153 (53%), Gaps = 2/153 (1%)
Frame = +3
Query: 62 GDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYGERNGKDNDMMPII 121
G + TV +AV + P+ +R V++I G Y E + + KR V F G+ GK +I
Sbjct: 3 GCYKTVQEAVNAAPANGTKRFVIYIKEGVYEETVRIPLEKRNVVFLGDGIGKT-----VI 167
Query: 122 TYDATALRYG--TLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALAMRISGDKAA 179
T + + G T +SATVAV D F+A + N++ PD + QA+A R+ D +
Sbjct: 168 TGNGNVGQQGMTTYNSATVAVLGDGFMAKELTVENTAG-PDAH----QAVAFRLDSDLSV 332
Query: 180 FYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDF 212
NC+F+G QDTL + F+K C I+G+ DF
Sbjct: 333 IENCEFLGNQDTLYAHSLRQFYKSCRIEGSVDF 431
>CA936291
Length = 429
Score = 82.0 bits (201), Expect = 3e-16
Identities = 53/133 (39%), Positives = 76/133 (56%), Gaps = 2/133 (1%)
Frame = +1
Query: 91 YREKITVDRSKRFVTFYGERNGKDNDMMPIITYDATALRYG--TLDSATVAVDADYFVAV 148
Y+E + V +KR V F G+ GK +IT DA + G T +SATVAV D F+A
Sbjct: 4 YQETVRVPLAKRNVVFLGDGIGKT-----VITGDANVGQQGMTTYNSATVAVLGDGFMAK 168
Query: 149 NVAFVNSSPMPDENSVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQG 208
++ N++ PD + QA+A R+ D + NC+F+G QDTL + F+K C I+G
Sbjct: 169 DLTIENTAG-PDAH----QAVAFRLDSDLSVIENCEFLGNQDTLYAHSLRQFYKSCRIEG 333
Query: 209 TYDFIFGNGKSIY 221
DFIFGN +I+
Sbjct: 334 NVDFIFGNAAAIF 372
>TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (64%)
Length = 1177
Score = 82.0 bits (201), Expect = 3e-16
Identities = 58/184 (31%), Positives = 97/184 (52%), Gaps = 1/184 (0%)
Frame = +3
Query: 57 RKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYGERNGKDND 116
R G ++T V DAV + P+ + +R V+ I G Y E + + + K + G+ G D
Sbjct: 198 RPTGRENYTKVMDAVLAAPNYSMQRYVIHIKRGVYYENVEIKKKKWNLMMVGD--GMD-- 365
Query: 117 MMPIITYDATALR-YGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALAMRISG 175
II+ + + + + T SAT AV F+A ++ F N++ P+++ QA+A+R
Sbjct: 366 -ATIISGNRSFIDGWTTFRSATFAVSGRGFIARDITFQNTAG-PEKH----QAVALRSDS 527
Query: 176 DKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIYLRTTIESVAKGFEC 235
D + F+ C G+QD+L + F+++C I GT DFIFG+ +I+ I S KG
Sbjct: 528 DLSVFFRCGIFGYQDSLYTHTMRQFYRECKISGTVDFIFGDATAIFQNCHI-SAKKGLPN 704
Query: 236 HHST 239
+T
Sbjct: 705 QKNT 716
>TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4L23.27
{Arabidopsis thaliana;} , partial (69%)
Length = 1483
Score = 73.2 bits (178), Expect(2) = 7e-16
Identities = 52/151 (34%), Positives = 78/151 (51%), Gaps = 2/151 (1%)
Frame = +2
Query: 54 VRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYGERNGK 113
V V KDG+G +TTV AV + P + R V+++ G Y E++ V + + G+ GK
Sbjct: 740 VVVAKDGSGKYTTVKAAVDAAPKSSSGRYVIYVKSGVYNEQVEVKGNN--IMLVGDGIGK 913
Query: 114 DNDMMPIITYDATALRYG--TLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALAM 171
IIT + ++ G T SATVA D F+A ++ F N++ + QA+A
Sbjct: 914 -----TIIT-GSKSVGGGTTTFRSATVAAVGDGFIAQDITFRNTA-----GAANHQAVAF 1060
Query: 172 RISGDKAAFYNCKFIGFQDTLCDDYGKHFFK 202
R D + FY C F GFQDTL + F++
Sbjct: 1061RSGSDLSVFYRCSFEGFQDTLYVHSERQFYR 1153
Score = 28.1 bits (61), Expect(2) = 7e-16
Identities = 18/41 (43%), Positives = 22/41 (52%), Gaps = 10/41 (24%)
Frame = +3
Query: 202 KDCFIQGTYDFIFGNGK------SIYLRT----TIESVAKG 232
++C I GT DFIFGN +IY RT TI A+G
Sbjct: 1152 EECDIYGTVDFIFGNAAAVLQNCNIYARTPPQRTITVTAQG 1274
>BQ628447 similar to GP|10178067|db pectin methylesterase-like protein
{Arabidopsis thaliana}, partial (22%)
Length = 446
Score = 79.7 bits (195), Expect = 2e-15
Identities = 53/149 (35%), Positives = 77/149 (51%), Gaps = 2/149 (1%)
Frame = +2
Query: 66 TVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYGERNGKDNDMMPIITYDA 125
TV +AV + P ++R V++I G Y E++ V KR V F G+ GK +IT A
Sbjct: 29 TVQEAVNAAPDEGEKRFVIYIKEGVYEERVRVPLKKRNVVFLGDGMGKT-----VITGSA 193
Query: 126 TALRYG--TLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALAMRISGDKAAFYNC 183
+ G T +SATV V D F+A ++ N++ + QA+A R D + NC
Sbjct: 194 NVGQPGMTTYNSATVGVAGDGFIAKDLTIQNTA-----GANAHQAVAFRSDSDLSVIENC 358
Query: 184 KFIGFQDTLCDDYGKHFFKDCFIQGTYDF 212
+FIG QDTL + F++ C I G DF
Sbjct: 359 EFIGNQDTLYAHSLRQFYRSCRIIGNVDF 445
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.328 0.141 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,257,416
Number of Sequences: 63676
Number of extensions: 292721
Number of successful extensions: 1944
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 1907
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1920
length of query: 335
length of database: 12,639,632
effective HSP length: 98
effective length of query: 237
effective length of database: 6,399,384
effective search space: 1516654008
effective search space used: 1516654008
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC124967.4


