

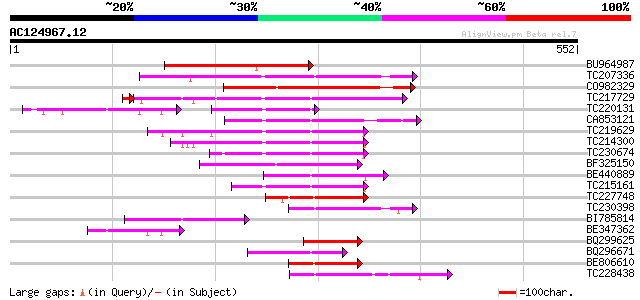
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124967.12 - phase: 0
(552 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BU964987 similar to GP|15208447|gb kinesin heavy chain {Zea mays... 161 8e-40
TC207336 similar to UP|O24147 (O24147) Kinesin-like protein, par... 146 3e-35
CO982329 144 1e-34
TC217729 similar to UP|Q9CAC9 (Q9CAC9) Kinesin-like protein; 736... 123 9e-29
TC220131 similar to UP|ATK2_ARATH (P46864) Kinesin 2 (Kinesin-li... 71 1e-26
CA853121 109 4e-24
TC219629 similar to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-li... 108 5e-24
TC214300 similar to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-li... 103 2e-22
TC230674 homologue to UP|Q9M0X6 (Q9M0X6) Kinesin-like protein, p... 96 4e-20
BF325150 weakly similar to GP|21741211|e OSJNBb0086G13.6 {Oryza ... 90 2e-18
BE440889 90 3e-18
TC215161 similar to UP|Q6NQ77 (Q6NQ77) At4g05190, partial (19%) 87 1e-17
TC227748 similar to UP|Q940Y8 (Q940Y8) AT3g16060/MSL1_10, partia... 83 3e-16
TC230398 homologue to UP|O24147 (O24147) Kinesin-like protein, p... 74 2e-13
BI785814 similar to GP|11761076|db kinesin-like protein {Oryza s... 74 2e-13
BE347362 69 4e-12
BQ299625 homologue to GP|21741211|e OSJNBb0086G13.6 {Oryza sativ... 67 3e-11
BQ296671 similar to GP|18087642|gb AT5g27950/F15F15_20 {Arabidop... 66 5e-11
BE806610 homologue to GP|15450501|gb| AT3g16060/MSL1_10 {Arabido... 64 2e-10
TC228438 similar to UP|Q6WJ05 (Q6WJ05) Central motor kinesin 1, ... 62 5e-10
>BU964987 similar to GP|15208447|gb kinesin heavy chain {Zea mays}, partial
(19%)
Length = 443
Score = 161 bits (407), Expect = 8e-40
Identities = 88/147 (59%), Positives = 105/147 (70%), Gaps = 2/147 (1%)
Frame = +2
Query: 151 KISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLRHLIGICEAHR 210
K SA+EIYNE + DLL+ E+ LR+ DD E+ +VE L EE R+ HL+ L+ CEA R
Sbjct: 2 KFSAIEIYNEIIRDLLSTENTSLRLRDDPERGPIVEKLTEETLRNWVHLKELLSFCEAQR 181
Query: 211 QVGETTLNDKSSRSHQIIRLTVESFHRE--SPDHVKSYIASLNFVDLAGSERASQTNTCG 268
QVGET LNDKSSRSHQIIRLT+ES RE + AS+NFVDLAGSERASQ + G
Sbjct: 182 QVGETYLNDKSSRSHQIIRLTIESSAREFMGKSSSTTLAASVNFVDLAGSERASQALSAG 361
Query: 269 TRLKEGSHINKSLLQLALVIRQLSSGK 295
RLKEG HIN+SLL L VIR+LS G+
Sbjct: 362 ARLKEGCHINRSLLTLGTVIRKLSKGR 442
>TC207336 similar to UP|O24147 (O24147) Kinesin-like protein, partial (22%)
Length = 1055
Score = 146 bits (368), Expect = 3e-35
Identities = 97/277 (35%), Positives = 159/277 (57%), Gaps = 6/277 (2%)
Frame = +3
Query: 127 GITENAIRDIYECIKNTPDR-DFVLKISALEIYNETVIDLLNRESG-PLR--ILDDTEKV 182
G+T AI +++ ++ ++ F LK +E+Y +T+IDLL ++G PL+ I D+ +
Sbjct: 9 GLTPRAIAELFRILRRDNNKYSFSLKAYMVELYQDTLIDLLLPKNGKPLKLDIKKDSTGM 188
Query: 183 TVVENLFEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDH 242
VVEN+ + L +I R + T +ND+SSRSH I+ + +ES + +S
Sbjct: 189 VVVENVTVMSISTIEELNSIIQRGSERRHISGTQMNDESSRSHLILSIVIESTNLQSQSV 368
Query: 243 VKSYIASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYR 302
K L+FVDLAGSER ++ + G++LKE INKSL L VI LSSG H YR
Sbjct: 369 AKG---KLSFVDLAGSERVKKSGSTGSQLKEAQSINKSLSALGDVISSLSSGGQ-HTPYR 536
Query: 303 TSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMVEGEL 362
KLT ++ SLGGNA+T + V+P+ S++++T N+L +A+ + ++N N+ E+
Sbjct: 537 NHKLTMLMSDSLGGNAKTLMFVNVAPTESNLDETNNSLMYASRVRSIVNDPNKNVSSKEV 716
Query: 363 RNPEPEHAGLRSLLAE-KELKIQQME-KDMEDLRRQR 397
A L+ L+A K+ + +E D+E+++ +R
Sbjct: 717 -------ARLKKLVAYWKQQAGRTLEYDDLEEIQDER 806
>CO982329
Length = 852
Score = 144 bits (363), Expect = 1e-34
Identities = 85/188 (45%), Positives = 119/188 (63%), Gaps = 1/188 (0%)
Frame = -3
Query: 209 HRQVGETTLNDKSSRSHQIIRLTVESFH-RESPDHVKSYIASLNFVDLAGSERASQTNTC 267
HR VG T N SSRSH I LT+ES E+ + ++ LN +DLAGSE +S+ T
Sbjct: 835 HRHVGSTNFNLLSSRSHTIFSLTIESSPCGENNEGEAITLSQLNLIDLAGSE-SSRAETT 659
Query: 268 GTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAIICTVS 327
G R +EGS+INKSLL L VI +L+ GK+ HI YR SKLTR+LQSSL G+ R ++ICTV+
Sbjct: 658 GMRRREGSYINKSLLTLGTVISKLTEGKASHIPYRDSKLTRLLQSSLSGHGRISLICTVT 479
Query: 328 PSLSHVEQTRNTLSFATNAKEVINTARVNMVEGELRNPEPEHAGLRSLLAEKELKIQQME 387
PS S+ E+T NTL FA AK + A N + E +SL+ + + +IQ ++
Sbjct: 478 PSSSNAEETHNTLKFAHRAKHIEIQAAQNTIIDE-----------KSLIKKYQHEIQFLK 332
Query: 388 KDMEDLRR 395
+++E +R+
Sbjct: 331 EELEQMRQ 308
>TC217729 similar to UP|Q9CAC9 (Q9CAC9) Kinesin-like protein; 73641-79546,
partial (26%)
Length = 1075
Score = 123 bits (309), Expect(2) = 9e-29
Identities = 93/284 (32%), Positives = 147/284 (51%), Gaps = 17/284 (5%)
Frame = +1
Query: 121 KTFTMRG----------ITENAIRDIYECIKNTPDRD-FVLKISALEIYNETVIDLLNRE 169
KT+TM G + A+ D++ ++ + + + +EIYNE V DLL+
Sbjct: 37 KTYTMSGPGLSSKSDWGVNYRALHDLFHISQSRRSSIVYEVGVQMVEIYNEQVRDLLS-S 213
Query: 170 SGPLRILD-----DTEKVTVVENLFEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRS 224
+GP + L + V + V A L L+ I +R T LN++SSRS
Sbjct: 214 NGPQKRLGIWNTAQPNGLAVPDASMHSVNSMADVLE-LMNIGLMNRATSATALNERSSRS 390
Query: 225 HQIIRLTVESFHRESPDHVKSYIASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQL 284
H ++ + V ++ ++ L+ VDLAGSER ++ G LKE HINKSL L
Sbjct: 391 HSVLSVHVRGTDLKTNTLLRG---CLHLVDLAGSERVDRSEATGDMLKEAQHINKSLSAL 561
Query: 285 ALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFAT 344
VI LS KS H+ YR SKLT++LQSSLGG A+T + ++P ++ +T +TL FA
Sbjct: 562 GDVIFALSQ-KSSHVPYRNSKLTQLLQSSLGGQAKTLMFVQLNPDVASYSETVSTLKFAE 738
Query: 345 NAKEV-INTARVNMVEGELRNPEPEHAGLRSLLAEKELKIQQME 387
V + AR N ++R + A L+ ++A K+ +I++++
Sbjct: 739 RVSGVELGAARSNKEGRDVRELMEQLASLKDVIARKDEEIERLQ 870
Score = 21.9 bits (45), Expect(2) = 9e-29
Identities = 9/11 (81%), Positives = 9/11 (81%)
Frame = +3
Query: 111 IFAYGQTSSGK 121
IFAYGQT S K
Sbjct: 6 IFAYGQTGSXK 38
>TC220131 similar to UP|ATK2_ARATH (P46864) Kinesin 2 (Kinesin-like protein
B), partial (37%)
Length = 1044
Score = 70.9 bits (172), Expect(2) = 1e-26
Identities = 54/170 (31%), Positives = 90/170 (52%), Gaps = 15/170 (8%)
Frame = +3
Query: 13 KIRRKLADTPGGSKIREEK--IRVTVRMRPLNRKEQAMYD--LIAWDCLDDKTIVFKNPN 68
++R+KL +T I E K IRV R+RPL E + + ++ + + +
Sbjct: 132 RLRKKLHNT-----ILELKGNIRVFCRVRPLLADESCSTEGRIFSYPTSMETSGRAIDLA 296
Query: 69 QERPSTSYTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTM--- 125
Q ++TFD+VF P S ++V+ E + SAL G IFAYGQT SGKT+TM
Sbjct: 297 QNGQKHAFTFDKVFTPEASQEEVFVE-ISQLVQSALDGYKVCIFAYGQTGSGKTYTMMGR 473
Query: 126 ------RGITENAIRDIYECIKNTPDR--DFVLKISALEIYNETVIDLLN 167
+G+ ++ I++ ++ + + +++S LEIYNET+ DL++
Sbjct: 474 PGHPEEKGLIPRSLEQIFQTKQSQQPQGWKYEMQVSMLEIYNETIRDLIS 623
Score = 67.8 bits (164), Expect(2) = 1e-26
Identities = 41/105 (39%), Positives = 59/105 (56%)
Frame = +1
Query: 197 QHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKSYIASLNFVDLA 256
Q L L+ R VG+T +N++SSRSH + L + + + V+ LN +DLA
Sbjct: 742 QRLHFLLNQAANSRSVGKTQMNEQSSRSHFVFTLRIYGVNESTDQQVQGV---LNLIDLA 912
Query: 257 GSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISY 301
GSER S++ + G RLKE INKSL L+ VI L+ K H+ +
Sbjct: 913 GSERLSKSGSTGDRLKETQAINKSLSSLSDVIFALAK-KEDHVPF 1044
>CA853121
Length = 677
Score = 109 bits (272), Expect = 4e-24
Identities = 75/192 (39%), Positives = 107/192 (55%)
Frame = +3
Query: 210 RQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKSYIASLNFVDLAGSERASQTNTCGT 269
R VG T+ N+ SSRSH ++R+TV + + +S+ L VDLAGSER +T G
Sbjct: 111 RSVGSTSANELSSRSHCLLRVTVLGENLINGQKTRSH---LWLVDLAGSERVGKTEAEGE 281
Query: 270 RLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAIICTVSPS 329
RLKE INKSL L VI L+S KS HI YR SKLT ILQSSLGG+ +T + +SP
Sbjct: 282 RLKESQFINKSLSALGDVISALAS-KSAHIPYRNSKLTHILQSSLGGDCKTLMFVQISPG 458
Query: 330 LSHVEQTRNTLSFATNAKEVINTARVNMVEGELRNPEPEHAGLRSLLAEKELKIQQMEKD 389
+ + +T +L+FAT V G P + L L K++ +++++ D
Sbjct: 459 AADLTETLCSLNFATR------------VRGIESGPARKQTDLTELNKYKQM-VEKVKHD 599
Query: 390 MEDLRRQRDLAQ 401
++ R+ +D Q
Sbjct: 600 EKETRKLQDNLQ 635
>TC219629 similar to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-like protein
A), partial (26%)
Length = 923
Score = 108 bits (271), Expect = 5e-24
Identities = 78/229 (34%), Positives = 120/229 (52%), Gaps = 14/229 (6%)
Frame = +1
Query: 135 DIYECIKNTPDRD--FVLKISALEIYNETVIDLL--NRESGPLRILDDTEKVTVVENLFE 190
+I++ ++ D+ + + +S EIYNET+ DLL NR SG + T +
Sbjct: 7 EIFQTSQSLKDQGWKYTMHVSIYEIYNETIRDLLSSNRSSGNDHTRTENSAPTPSKQHTI 186
Query: 191 EVARD--------AQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDH 242
+ D A+ + L+ R VG T +N++SSRSH + +L + + ++
Sbjct: 187 KHESDLATLEVCSAEEISSLLQQAAQSRSVGRTQMNEQSSRSHFVFKLRISGRNEKTEKQ 366
Query: 243 VKSYIASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYR 302
V+ LN +DLAGSER S++ G RLKE INKSL L+ VI L+ K H+ +R
Sbjct: 367 VQGV---LNLIDLAGSERLSRSGATGDRLKETQAINKSLSSLSDVIFALAK-KEEHVPFR 534
Query: 303 TSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFA--TNAKEV 349
SKLT LQ LGG+++T + +SP S ++ +L FA NA E+
Sbjct: 535 NSKLTHFLQPYLGGDSKTLMFVNISPDQSSAGESLCSLRFAARVNACEI 681
>TC214300 similar to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-like protein
A), partial (27%)
Length = 903
Score = 103 bits (257), Expect = 2e-22
Identities = 75/211 (35%), Positives = 113/211 (53%), Gaps = 18/211 (8%)
Frame = +2
Query: 157 IYNETVIDLL--NRESG---PLRILD-----------DTEKVTVVENLFEEVARDAQHLR 200
IYNET+ DLL N+ S P R+ + D T V +L + + +
Sbjct: 2 IYNETIRDLLATNKSSADGTPTRVENGTPGKQYMIKHDANGNTHVSDLTVVDVQSVKEVA 181
Query: 201 HLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKSYIASLNFVDLAGSER 260
L+ + R VG+T +N++SSRSH + L + + + V+ LN +DLAGSER
Sbjct: 182 FLLNQAASSRSVGKTQMNEQSSRSHFVFTLRIYGVNESTDQQVQGI---LNLIDLAGSER 352
Query: 261 ASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNART 320
S++ + G RLKE INKSL L+ VI L+ K HI +R SKLT +LQ LGG+++T
Sbjct: 353 LSRSGSTGDRLKETQAINKSLSSLSDVIFALAK-KEDHIPFRNSKLTYLLQPCLGGDSKT 529
Query: 321 AIICTVSPSLSHVEQTRNTLSFAT--NAKEV 349
+ +SP + ++ +L FA+ NA E+
Sbjct: 530 LMFVNISPDQASSGESLCSLRFASRVNACEI 622
>TC230674 homologue to UP|Q9M0X6 (Q9M0X6) Kinesin-like protein, partial (20%)
Length = 811
Score = 95.9 bits (237), Expect = 4e-20
Identities = 61/157 (38%), Positives = 89/157 (55%), Gaps = 2/157 (1%)
Frame = +1
Query: 195 DAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKSYIASLNFVD 254
D ++R + C R VG T +N++SSRSH + L + + + V+ LN +D
Sbjct: 55 DXXYIRFNVPSCS--RSVGRTHMNEQSSRSHFVXTLRISGTNSNTDQQVQGV---LNLID 219
Query: 255 LAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSL 314
LAGSER S++ G RLKE INKSL L+ VI L+ K H+ +R SKLT +LQ L
Sbjct: 220 LAGSERLSRSGATGDRLKETQAINKSLSSLSDVIFALAK-KQEHVPFRNSKLTYLLQPCL 396
Query: 315 GGNARTAIICTVSPSLSHVEQTRNTLSFA--TNAKEV 349
GG+++T + +SP S ++ +L FA NA E+
Sbjct: 397 GGDSKTLMFVNISPDPSSTGESLCSLRFAAGVNACEI 507
>BF325150 weakly similar to GP|21741211|e OSJNBb0086G13.6 {Oryza sativa
(japonica cultivar-group)}, partial (12%)
Length = 547
Score = 90.1 bits (222), Expect = 2e-18
Identities = 54/160 (33%), Positives = 91/160 (56%), Gaps = 1/160 (0%)
Frame = +2
Query: 185 VENLFEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVE-SFHRESPDHV 243
++ + EEV H LI E H + T N S+ SH I LT+E S + ++ +
Sbjct: 5 IKGIKEEVILSPTHTLSLIATEEEHXHIKSTNFNLLSTESHTIFALTIENSPYNKNNERE 184
Query: 244 KSYIASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRT 303
++ LN ++L S ++S+T T R ++ S+INK+LL L +I +L+ ++ I Y+
Sbjct: 185 XVTLSQLNLINLTNS-KSSKTKTTNIRKRQRSYINKTLLTLKTIISKLT*NRATRIPYKN 361
Query: 304 SKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFA 343
SKLT++L SSL + +++ TV+PS S + T NTL+ +
Sbjct: 362 SKLTKLLDSSLRKHKHISLMYTVTPSSSDTKYTHNTLNLS 481
>BE440889
Length = 484
Score = 89.7 bits (221), Expect = 3e-18
Identities = 55/129 (42%), Positives = 78/129 (59%), Gaps = 8/129 (6%)
Frame = +2
Query: 248 ASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLT 307
+ L VDLAGSER ++T+ G RLKE +IN+SL L VI L++ KS HI YR SKLT
Sbjct: 83 SKLWLVDLAGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAA-KSSHIPYRNSKLT 259
Query: 308 RILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATN--------AKEVINTARVNMVE 359
+LQ SLGG+++T + +SPS V +T ++L+FAT K+ I+T+ V ++
Sbjct: 260 HLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELGPVKKQIDTSEVQKMK 439
Query: 360 GELRNPEPE 368
L E
Sbjct: 440 AMLEKARSE 466
>TC215161 similar to UP|Q6NQ77 (Q6NQ77) At4g05190, partial (19%)
Length = 692
Score = 87.4 bits (215), Expect = 1e-17
Identities = 55/135 (40%), Positives = 80/135 (58%), Gaps = 2/135 (1%)
Frame = +2
Query: 217 LNDKSSRSHQIIRLTVESFHRESPDHVKSYIASLNFVDLAGSERASQTNTCGTRLKEGSH 276
+N++SSRSH + L + + + V+ LN +DLAGSER S++ + G RLKE
Sbjct: 41 MNEQSSRSHFVFTLRIYGVNESTDXXVQGV---LNLIDLAGSERLSKSGSTGDRLKETQA 211
Query: 277 INKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAIICTVSPSLSHVEQT 336
INKSL L+ VI L+ K H+ +R SKLT +LQ LGG + T + +SP S V ++
Sbjct: 212 INKSLSSLSDVIFALAK-KEDHVPFRNSKLTYLLQPCLGGYSNTLMFVNISPDPSSVGES 388
Query: 337 RNTLSFAT--NAKEV 349
+L FA+ NA E+
Sbjct: 389 LCSLRFASRVNACEI 433
>TC227748 similar to UP|Q940Y8 (Q940Y8) AT3g16060/MSL1_10, partial (34%)
Length = 1192
Score = 83.2 bits (204), Expect = 3e-16
Identities = 50/102 (49%), Positives = 65/102 (63%), Gaps = 2/102 (1%)
Frame = +3
Query: 250 LNFVDLAGSERASQT--NTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLT 307
L+F+DLAGSER + T N TR+ EG+ INKSLL L IR L + + GHI +R SKLT
Sbjct: 6 LSFIDLAGSERGADTTDNDKQTRI-EGAEINKSLLALKECIRALDNDQ-GHIPFRGSKLT 179
Query: 308 RILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEV 349
+L+ S GN+RT +I +SPS E T NTL +A K +
Sbjct: 180 EVLRDSFVGNSRTVMISCISPSTGSCEHTLNTLRYADRVKSL 305
>TC230398 homologue to UP|O24147 (O24147) Kinesin-like protein, partial (10%)
Length = 599
Score = 73.9 bits (180), Expect = 2e-13
Identities = 45/128 (35%), Positives = 73/128 (56%), Gaps = 2/128 (1%)
Frame = +3
Query: 272 KEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAIICTVSPSLS 331
KE INKSL L VI LSSG HI YR KLT ++ SLGGNA+T + VSP S
Sbjct: 3 KEAQSINKSLSALGDVISALSSGGQ-HIPYRNHKLTMLMSDSLGGNAKTLMFVNVSPVES 179
Query: 332 HVEQTRNTLSFATNAKEVINTARVNMVEGELRNPEPEHAGLRSLLA--EKELKIQQMEKD 389
+++T N+L +A+ + ++N N+ E+ A L+ L+ +++ + ++D
Sbjct: 180 SLDETHNSLMYASRVRSIVNDPSKNVSSKEI-------ARLKKLIGYWKEQAGRRGEDED 338
Query: 390 MEDLRRQR 397
+E+++ +R
Sbjct: 339 LEEIQEER 362
>BI785814 similar to GP|11761076|db kinesin-like protein {Oryza sativa
(japonica cultivar-group)}, partial (19%)
Length = 421
Score = 73.6 bits (179), Expect = 2e-13
Identities = 45/123 (36%), Positives = 71/123 (57%), Gaps = 1/123 (0%)
Frame = +3
Query: 112 FAYGQTSSGKTFTMRGITENAIRDIYECIKNT-PDRDFVLKISALEIYNETVIDLLNRES 170
FAYGQT SGKT+TM+ + A RDI + +T ++ F L +S EIY + DLLN +
Sbjct: 6 FAYGQTGSGKTYTMKPLPLKASRDILRLMHHTYRNQGFQLFVSFFEIYGGKLFDLLN-DR 182
Query: 171 GPLRILDDTEKVTVVENLFEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRL 230
L + +D ++ + L E D ++++ LI + R G T N++SSRSH I++L
Sbjct: 183 KKLCMREDGKQQVCIVGLQEYRVSDVENIKDLIEKGNSTRSTGTTGANEESSRSHAILQL 362
Query: 231 TVE 233
++
Sbjct: 363 AIK 371
>BE347362
Length = 448
Score = 69.3 bits (168), Expect = 4e-12
Identities = 41/102 (40%), Positives = 56/102 (54%), Gaps = 7/102 (6%)
Frame = +1
Query: 76 YTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENA--- 132
+ FD VF P + + V+ + K + S L G N IFAYGQT +GKTFTM G E+
Sbjct: 64 FKFDHVFGPEDNQETVFQQ-TKPIVTSVLDGYNVCIFAYGQTGTGKTFTMEGTPEHRGVN 240
Query: 133 IRDIYECIKNTPDR----DFVLKISALEIYNETVIDLLNRES 170
R + E + T +R + L +S LE+YNE + DLL S
Sbjct: 241 YRTLEELFRITEERHGTMKYELSVSMLEVYNEKIRDLLVENS 366
>BQ299625 homologue to GP|21741211|e OSJNBb0086G13.6 {Oryza sativa (japonica
cultivar-group)}, partial (9%)
Length = 406
Score = 66.6 bits (161), Expect = 3e-11
Identities = 33/57 (57%), Positives = 42/57 (72%)
Frame = +2
Query: 287 VIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFA 343
VI +L+ GK HI YR SKLTR+LQSSL G+ R ++ICTV+ + S E+T NTL FA
Sbjct: 119 VIAKLTDGKETHIPYRDSKLTRLLQSSLSGHGRISLICTVTLASSSSEETHNTLKFA 289
>BQ296671 similar to GP|18087642|gb AT5g27950/F15F15_20 {Arabidopsis
thaliana}, partial (20%)
Length = 422
Score = 65.9 bits (159), Expect = 5e-11
Identities = 40/98 (40%), Positives = 54/98 (54%)
Frame = +3
Query: 232 VESFHRESPDHVKSYIASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQL 291
+ F R KS ++ L +DL GSER +T G L EG IN SL LA V+ L
Sbjct: 114 ISIFRRGDALEAKSEVSKLWMIDLGGSERLLKTGAKGLTLDEGRAINLSLSALADVVAAL 293
Query: 292 SSGKSGHISYRTSKLTRILQSSLGGNARTAIICTVSPS 329
K H+ YR SKLT+IL+ SLG ++ ++ +SPS
Sbjct: 294 KR-KRCHVPYRNSKLTQILKDSLGYGSKVLMLVHISPS 404
>BE806610 homologue to GP|15450501|gb| AT3g16060/MSL1_10 {Arabidopsis
thaliana}, partial (10%)
Length = 220
Score = 63.9 bits (154), Expect = 2e-10
Identities = 35/72 (48%), Positives = 46/72 (63%)
Frame = +2
Query: 272 KEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAIICTVSPSLS 331
+EG+ INKSLL L IR L + + GHI +R SKLT +L+ S GN+RT +I +SP
Sbjct: 2 QEGAEINKSLLVLKECIRALDNDQ-GHIPFRGSKLTEVLRDSFVGNSRTVMISCISPMFG 178
Query: 332 HVEQTRNTLSFA 343
E T NTL +A
Sbjct: 179 SCEHTLNTLRYA 214
>TC228438 similar to UP|Q6WJ05 (Q6WJ05) Central motor kinesin 1, partial
(13%)
Length = 809
Score = 62.4 bits (150), Expect = 5e-10
Identities = 48/161 (29%), Positives = 77/161 (47%), Gaps = 2/161 (1%)
Frame = +2
Query: 273 EGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAIICTVSPSLSH 332
EG+ INKSLL L IR L + + HI +R SKLT +L+ S GN++T +I +SP
Sbjct: 5 EGA*INKSLLALKECIRALDNDQI-HIPFRGSKLTEVLRDSFVGNSKTVMISCISPGAGS 181
Query: 333 VEQTRNTLSFATNAKEVINTARVNMVEGELRNPEPEHAGLRSLLAEKELKIQQMEKDMED 392
E T NTL +A K + + N + ++ N P+ + + + +D D
Sbjct: 182 CEHTLNTLRYADRVKSLSKSG--NPRKDQVPNAVPQ-TNNKDVSSTSSFPASSGAEDFND 352
Query: 393 LRRQR--DLAQCQLDLERRANKVQKGSSDYGPSSQVVRCLS 431
R+++ D+ + ++ E + S D P S LS
Sbjct: 353 QRQEKTIDMGRKFVEKENSLHSSAAASVDKQPVSYSSNYLS 475
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.130 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,421,311
Number of Sequences: 63676
Number of extensions: 232335
Number of successful extensions: 1131
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 1099
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1103
length of query: 552
length of database: 12,639,632
effective HSP length: 102
effective length of query: 450
effective length of database: 6,144,680
effective search space: 2765106000
effective search space used: 2765106000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC124967.12


