

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124966.9 - phase: 0 /pseudo
(86 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
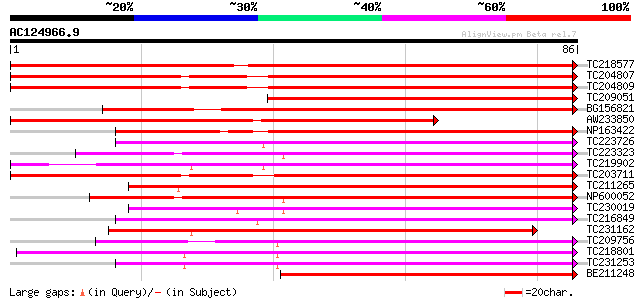
Score E
Sequences producing significant alignments: (bits) Value
TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (F... 102 3e-23
TC204807 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone g... 81 8e-17
TC204809 weakly similar to PIR|D86144|D86144 protein probable UT... 79 3e-16
TC209051 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferas... 79 4e-16
BG156821 similar to SP|Q9AR73|HQGT Hydroquinone glucosyltransfer... 77 2e-15
AW233850 similar to GP|19911209|db glucosyltransferase-13 {Vigna... 76 3e-15
NP163422 UDP-glycose:flavonoid glycosyltransferase 75 4e-15
TC223726 weakly similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransf... 67 1e-12
TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransfer... 63 2e-11
TC219902 similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransferase H... 63 2e-11
TC203711 similar to UP|Q8S9A4 (Q8S9A4) Glucosyltransferase-5, pa... 63 3e-11
TC211265 similar to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid gly... 61 1e-10
NP600052 zeatin O-glucosyltransferase [Glycine max] 60 2e-10
TC230019 weakly similar to UP|Q6WFW1 (Q6WFW1) Glucosyltransferas... 59 3e-10
TC216849 weakly similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransf... 59 4e-10
TC231162 weakly similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransf... 59 4e-10
TC209756 56 3e-09
TC218801 similar to UP|Q8W3P8 (Q8W3P8) ABA-glucosyltransferase, ... 55 6e-09
TC231253 weakly similar to UP|Q8S997 (Q8S997) Glucosyltransferas... 55 8e-09
BE211248 similar to SP|Q9ZSK5|ZOG_ Zeatin O-glucosyltransferase ... 54 1e-08
>TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (Fragment),
partial (58%)
Length = 1530
Score = 102 bits (255), Expect = 3e-23
Identities = 52/86 (60%), Positives = 61/86 (70%)
Frame = +1
Query: 1 MEMGPIKAPTEEGSGNPSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGS 60
+E G I+A E G P +YPVGPII + NG+ECL+WLDKQ+ SVLYVSFGS
Sbjct: 634 LESGAIRALEEHVKGKPKLYPVGPIIQMESIGHE--NGVECLTWLDKQEPNSVLYVSFGS 807
Query: 61 GGTLSHEQIVQLALGLELSNQKFLWV 86
GGTLS EQ +LA GLELS +KFLWV
Sbjct: 808 GGTLSQEQFNELAFGLELSGKKFLWV 885
>TC204807 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone
glucosyltransferase (Arbutin synthase) , partial (79%)
Length = 1673
Score = 81.3 bits (199), Expect = 8e-17
Identities = 45/86 (52%), Positives = 55/86 (63%)
Frame = +2
Query: 1 MEMGPIKAPTEEGSGNPSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGS 60
+E G +E G P VY VGP++ + D+ ECL WLD+Q SVL+VSFGS
Sbjct: 893 LEPGAWNELQKEEQGRPPVYAVGPLV-RMEAGQADS---ECLRWLDEQPRGSVLFVSFGS 1060
Query: 61 GGTLSHEQIVQLALGLELSNQKFLWV 86
GGTLS QI +LALGLE S Q+FLWV
Sbjct: 1061GGTLSSAQINELALGLEKSEQRFLWV 1138
>TC204809 weakly similar to PIR|D86144|D86144 protein probable UTP-glucose
glucosyltransferase [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (23%)
Length = 807
Score = 79.3 bits (194), Expect = 3e-16
Identities = 45/86 (52%), Positives = 54/86 (62%)
Frame = +3
Query: 1 MEMGPIKAPTEEGSGNPSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGS 60
+E G +E G P VY VGP++ + D+ ECL WLD+Q SVL VSFGS
Sbjct: 261 LEPGAWNELQKEEQGRPPVYAVGPLV-RMEAGQADS---ECLRWLDEQPRGSVLCVSFGS 428
Query: 61 GGTLSHEQIVQLALGLELSNQKFLWV 86
GGTLS QI +LALGLE S Q+FLWV
Sbjct: 429 GGTLSSAQINELALGLEKSEQRFLWV 506
>TC209051 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13
(Fragment), partial (35%)
Length = 1017
Score = 79.0 bits (193), Expect = 4e-16
Identities = 37/47 (78%), Positives = 40/47 (84%)
Frame = +2
Query: 40 ECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGLELSNQKFLWV 86
+CLSWLDKQ CSVLYVSFGSGGTLS QI +LA GLELS Q+FLWV
Sbjct: 8 KCLSWLDKQPPCSVLYVSFGSGGTLSQNQINELASGLELSGQRFLWV 148
>BG156821 similar to SP|Q9AR73|HQGT Hydroquinone glucosyltransferase (EC
2.4.1.218) (Arbutin synthase). [Serpentwood
Devilpepper], partial (27%)
Length = 465
Score = 77.0 bits (188), Expect = 2e-15
Identities = 41/72 (56%), Positives = 48/72 (65%)
Frame = +2
Query: 15 GNPSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLAL 74
G P VY VGP++ + ECL WLD+Q SVL+VSFGSGGTLS QI +LAL
Sbjct: 14 GRPPVYAVGPLVRM----EPGPADSECLRWLDEQPRGSVLFVSFGSGGTLSSAQINELAL 181
Query: 75 GLELSNQKFLWV 86
GLE S Q+FLWV
Sbjct: 182GLENSQQRFLWV 217
>AW233850 similar to GP|19911209|db glucosyltransferase-13 {Vigna angularis},
partial (21%)
Length = 411
Score = 76.3 bits (186), Expect = 3e-15
Identities = 40/65 (61%), Positives = 46/65 (70%)
Frame = +1
Query: 1 MEMGPIKAPTEEGSGNPSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGS 60
ME PI+A +E +G P VYP+GPII T SD L+C+ WLDKQQ SVLYVSFGS
Sbjct: 217 MEKEPIRALAKEWNGYPPVYPIGPIIQTGIESDGPIE-LDCIKWLDKQQPKSVLYVSFGS 393
Query: 61 GGTLS 65
GGTLS
Sbjct: 394 GGTLS 408
>NP163422 UDP-glycose:flavonoid glycosyltransferase
Length = 731
Score = 75.5 bits (184), Expect = 4e-15
Identities = 38/70 (54%), Positives = 50/70 (71%)
Frame = +1
Query: 17 PSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGL 76
P V+ +GP+I + +C RD N ECLSWLD Q S SVL++SFGS G S Q+ ++A+GL
Sbjct: 349 PKVFCIGPVIASASCR-RDDN--ECLSWLDSQPSHSVLFLSFGSMGRFSRTQLGEIAIGL 519
Query: 77 ELSNQKFLWV 86
E S Q+FLWV
Sbjct: 520 EKSEQRFLWV 549
>TC223726 weakly similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransferase HRA25,
partial (20%)
Length = 412
Score = 67.4 bits (163), Expect = 1e-12
Identities = 32/74 (43%), Positives = 44/74 (59%), Gaps = 4/74 (5%)
Frame = +1
Query: 17 PSVYPVGPIIDTVTCSDRDAN----GLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQL 72
P + P+GP++ + N L CL WLD+ CSV+YV+FGS T S Q +L
Sbjct: 112 PQIIPIGPLLSSNHLRHSAGNFWPQDLTCLKWLDQHSPCSVIYVAFGSFTTFSPTQFQEL 291
Query: 73 ALGLELSNQKFLWV 86
LGLEL+N+ F+WV
Sbjct: 292 CLGLELTNRPFIWV 333
>TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransferase, partial
(89%)
Length = 1386
Score = 63.2 bits (152), Expect = 2e-11
Identities = 34/77 (44%), Positives = 46/77 (59%), Gaps = 1/77 (1%)
Frame = +1
Query: 11 EEGSGNPSVYPVGPIIDTVTCSDRDANGLE-CLSWLDKQQSCSVLYVSFGSGGTLSHEQI 69
E +G ++ +GP + + +D+ CL WLDKQ SVLYVSFG+ T EQI
Sbjct: 667 ERFTGGKKLWALGPF-NPLAFEKKDSKERHFCLEWLDKQDPNSVLYVSFGTTTTFKEEQI 843
Query: 70 VQLALGLELSNQKFLWV 86
++A GLE S QKF+WV
Sbjct: 844 KKIATGLEQSKQKFIWV 894
>TC219902 similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransferase HRA25,
partial (52%)
Length = 967
Score = 63.2 bits (152), Expect = 2e-11
Identities = 36/94 (38%), Positives = 53/94 (56%), Gaps = 8/94 (8%)
Frame = +1
Query: 1 MEMGPIKAPTEEGSGNPSVYPVGPII----DTVTCSDRDAN----GLECLSWLDKQQSCS 52
+E GP+ S P++ P+GP++ DT+ + L C+SWLD+Q S
Sbjct: 58 LEPGPL-------SSIPNLVPIGPLLRSYGDTIATAKSIGQYWEEDLSCMSWLDQQPHGS 216
Query: 53 VLYVSFGSGGTLSHEQIVQLALGLELSNQKFLWV 86
VLYV+FGS Q +LALG++L+N+ FLWV
Sbjct: 217 VLYVAFGSFTHFDQNQFNELALGIDLTNRPFLWV 318
>TC203711 similar to UP|Q8S9A4 (Q8S9A4) Glucosyltransferase-5, partial (57%)
Length = 1322
Score = 62.8 bits (151), Expect = 3e-11
Identities = 35/86 (40%), Positives = 53/86 (60%)
Frame = +3
Query: 1 MEMGPIKAPTEEGSGNPSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGS 60
+E I+A +E+ + P ++ VGP+I + + D CLSWL+ Q S SV+ + FGS
Sbjct: 363 IEEEAIRALSEDATVPPPLFCVGPVI-SAPYGEEDKG---CLSWLNLQPSQSVVLLCFGS 530
Query: 61 GGTLSHEQIVQLALGLELSNQKFLWV 86
G S Q+ ++A+GLE S Q+FLWV
Sbjct: 531 MGRFSRAQLKEIAIGLEKSEQRFLWV 608
>TC211265 similar to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid
glycosyltransferase, partial (41%)
Length = 616
Score = 60.8 bits (146), Expect = 1e-10
Identities = 33/72 (45%), Positives = 44/72 (60%), Gaps = 4/72 (5%)
Frame = +3
Query: 19 VYPVGP----IIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLAL 74
V+ VGP + TV S D + +CL+WLD ++ SVLY+ FGS +S EQ+ Q+A
Sbjct: 363 VWHVGPSSLMVQKTVKSSTVDESRHDCLTWLDSKKESSVLYICFGSLSLISDEQLYQIAT 542
Query: 75 GLELSNQKFLWV 86
GLE S FLWV
Sbjct: 543 GLEGSGHCFLWV 578
>NP600052 zeatin O-glucosyltransferase [Glycine max]
Length = 1395
Score = 59.7 bits (143), Expect = 2e-10
Identities = 30/75 (40%), Positives = 46/75 (61%), Gaps = 1/75 (1%)
Frame = +1
Query: 13 GSGNPSVYPVGPIIDTVTCSDRDANGLE-CLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQ 71
G ++ +GP + +T +D C+ WLDKQ++ SV+YVSFG+ + + Q Q
Sbjct: 682 GGSKKRLWALGPF-NPLTIEKKDPKTRHICIEWLDKQEANSVMYVSFGTTTSFTVAQFEQ 858
Query: 72 LALGLELSNQKFLWV 86
+A+GLE S QKF+WV
Sbjct: 859 IAIGLEQSKQKFIWV 903
>TC230019 weakly similar to UP|Q6WFW1 (Q6WFW1) Glucosyltransferase-like
protein , partial (36%)
Length = 1202
Score = 59.3 bits (142), Expect = 3e-10
Identities = 30/76 (39%), Positives = 45/76 (58%), Gaps = 8/76 (10%)
Frame = +2
Query: 19 VYPVGPIIDTVTCSD-RDANGLE-------CLSWLDKQQSCSVLYVSFGSGGTLSHEQIV 70
V+PVGP++ + D + G E C+ WLD + SVLY+SFGS T++ Q++
Sbjct: 395 VWPVGPLLPPASLMDSKHRAGKESGIALDACMQWLDSKDESSVLYISFGSQNTITASQMM 574
Query: 71 QLALGLELSNQKFLWV 86
LA GLE S + F+W+
Sbjct: 575 ALAEGLEESGRSFIWI 622
>TC216849 weakly similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransferase 85A8,
partial (47%)
Length = 1125
Score = 58.9 bits (141), Expect = 4e-10
Identities = 31/80 (38%), Positives = 46/80 (56%), Gaps = 10/80 (12%)
Frame = +2
Query: 17 PSVYPVGPIIDTVTCSDRDA----------NGLECLSWLDKQQSCSVLYVSFGSGGTLSH 66
PS+YP+GP + S + LECL WL+ ++S SV+YV+FGS +S
Sbjct: 233 PSLYPIGPFPLLLNQSPQSHLTSLGSNLWNEDLECLEWLESKESRSVVYVNFGSITVMSA 412
Query: 67 EQIVQLALGLELSNQKFLWV 86
EQ+++ A GL S + FLW+
Sbjct: 413 EQLLEFAWGLANSKKPFLWI 472
>TC231162 weakly similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransferase HRA25,
partial (35%)
Length = 912
Score = 58.9 bits (141), Expect = 4e-10
Identities = 28/69 (40%), Positives = 42/69 (60%), Gaps = 4/69 (5%)
Frame = +2
Query: 16 NPSVYPVGPII----DTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQ 71
+P + P+GP+I D + + CL+WLD+Q CSV+YV+FGS Q+ +
Sbjct: 680 SPKILPIGPLIGSGNDIRSLGQFWEEDVSCLTWLDQQPPCSVIYVAFGSSTIFDPHQLKE 859
Query: 72 LALGLELSN 80
LALGL+L+N
Sbjct: 860 LALGLDLTN 886
>TC209756
Length = 1356
Score = 56.2 bits (134), Expect = 3e-09
Identities = 30/78 (38%), Positives = 44/78 (55%), Gaps = 5/78 (6%)
Frame = +1
Query: 14 SGNPSVYPVGPIIDTVTCSDRDANGL-----ECLSWLDKQQSCSVLYVSFGSGGTLSHEQ 68
S +P P+GP+++ SD + CL WLD+Q SV+YVSFGS + Q
Sbjct: 220 SVSPKFLPIGPLME----SDNSKSAFWEEDTTCLEWLDQQPPQSVIYVSFGSLAVMDPNQ 387
Query: 69 IVQLALGLELSNQKFLWV 86
+LAL L+L ++ F+WV
Sbjct: 388 FKELALALDLLDKPFIWV 441
>TC218801 similar to UP|Q8W3P8 (Q8W3P8) ABA-glucosyltransferase, partial
(60%)
Length = 1281
Score = 55.1 bits (131), Expect = 6e-09
Identities = 32/93 (34%), Positives = 49/93 (52%), Gaps = 8/93 (8%)
Frame = +1
Query: 2 EMGPIKAPTEEGSGNPSVYPVGPI-IDTVTCSDRDANGL-------ECLSWLDKQQSCSV 53
++ P A + + +GP+ + T D+ G +CL+WL+ ++ SV
Sbjct: 112 DLEPAYAEQVKNKWGKKAWIIGPVSLCNRTAEDKTERGKLPTIDEEKCLNWLNSKKPNSV 291
Query: 54 LYVSFGSGGTLSHEQIVQLALGLELSNQKFLWV 86
LYVSFGS L EQ+ ++A GLE S Q F+WV
Sbjct: 292 LYVSFGSLLRLPSEQLKEIACGLEASEQSFIWV 390
>TC231253 weakly similar to UP|Q8S997 (Q8S997) Glucosyltransferase-12,
partial (31%)
Length = 841
Score = 54.7 bits (130), Expect = 8e-09
Identities = 33/83 (39%), Positives = 47/83 (55%), Gaps = 13/83 (15%)
Frame = +3
Query: 17 PSVYPVGPI-------IDTVTCSDRDANGL------ECLSWLDKQQSCSVLYVSFGSGGT 63
P VY +GP+ I T + S +G C++WLD Q++ SVLYVSFG+
Sbjct: 114 PKVYSIGPLHTLCKTMITTNSTSSPHKDGRLRKEDRSCITWLDHQKAKSVLYVSFGTVVN 293
Query: 64 LSHEQIVQLALGLELSNQKFLWV 86
LS+EQ+++ GL S + FLWV
Sbjct: 294 LSYEQLMEFWHGLVNSLKPFLWV 362
>BE211248 similar to SP|Q9ZSK5|ZOG_ Zeatin O-glucosyltransferase (EC
2.4.1.203) (Trans-zeatin O-beta-D-
glucosyltransferase)., partial (24%)
Length = 426
Score = 54.3 bits (129), Expect = 1e-08
Identities = 26/45 (57%), Positives = 33/45 (72%)
Frame = +2
Query: 42 LSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGLELSNQKFLWV 86
+ WLDKQ++ SVLYVS G+ S EQI ++A GLE S QKF+WV
Sbjct: 56 VEWLDKQEA*SVLYVSLGTTTCFSDEQIKEVANGLEKSKQKFIWV 190
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.135 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,525,918
Number of Sequences: 63676
Number of extensions: 63170
Number of successful extensions: 288
Number of sequences better than 10.0: 103
Number of HSP's better than 10.0 without gapping: 278
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 279
length of query: 86
length of database: 12,639,632
effective HSP length: 62
effective length of query: 24
effective length of database: 8,691,720
effective search space: 208601280
effective search space used: 208601280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 52 (24.6 bits)
Medicago: description of AC124966.9


