

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124966.6 + phase: 0
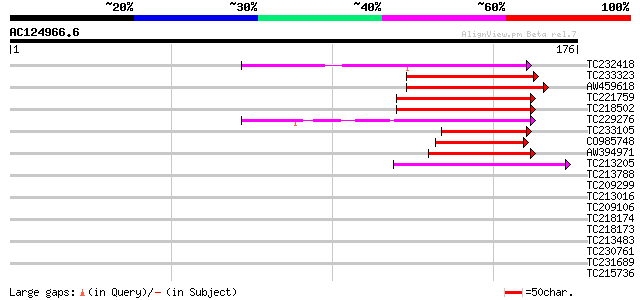
(176 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC232418 weakly similar to UP|O81348 (O81348) Symbiotic ammonium... 71 2e-13
TC233323 weakly similar to UP|O81348 (O81348) Symbiotic ammonium... 61 2e-10
AW459618 58 3e-09
TC221759 homologue to UP|O81348 (O81348) Symbiotic ammonium tran... 53 8e-08
TC218502 53 8e-08
TC229276 weakly similar to UP|SRE2_HUMAN (Q12772) Sterol regulat... 51 3e-07
TC233105 49 9e-07
CO985748 49 9e-07
AW394971 48 3e-06
TC213205 40 7e-04
TC213788 similar to GB|AAN73298.1|25141207|BT002301 At4g16430/dl... 37 0.005
TC209299 similar to GB|AAN73298.1|25141207|BT002301 At4g16430/dl... 37 0.005
TC213016 homologue to GB|AAN73298.1|25141207|BT002301 At4g16430/... 36 0.008
TC209106 similar to UP|Q41101 (Q41101) Phaseolin G-box binding p... 36 0.011
TC218174 similar to UP|Q41102 (Q41102) Phaseolin G-box binding p... 35 0.014
TC218173 similar to UP|Q41102 (Q41102) Phaseolin G-box binding p... 35 0.014
TC213483 similar to UP|Q8LPT0 (Q8LPT0) At2g46510/F13A10.4, parti... 35 0.018
TC230761 weakly similar to UP|O81348 (O81348) Symbiotic ammonium... 34 0.031
TC231689 weakly similar to UP|Q38736 (Q38736) DEL, partial (22%) 34 0.041
TC215736 similar to GB|AAO63377.1|28950907|BT005313 At4g34530 {A... 34 0.041
>TC232418 weakly similar to UP|O81348 (O81348) Symbiotic ammonium
transporter, partial (17%)
Length = 890
Score = 71.2 bits (173), Expect = 2e-13
Identities = 44/94 (46%), Positives = 56/94 (58%), Gaps = 4/94 (4%)
Frame = +2
Query: 73 PSQYILSFENSTMQPSPNSDIATCSSIMVQETTTLNNNVSSELPKIIKKR----TKNLRS 128
P YILSF+NSTM P+ S+ + L + E P + +R K +R+
Sbjct: 71 PRTYILSFDNSTMVPATPETRPRSSN-----NSPLPAKRALESPGPVARRPNQGAKKIRT 235
Query: 129 SCEMQDHIMAERKRRQVLTERFIALSATIPGLKK 162
S + DH+MAER+RRQ LTERFIALSATIPGL K
Sbjct: 236 SSQTIDHVMAERRRRQELTERFIALSATIPGLNK 337
>TC233323 weakly similar to UP|O81348 (O81348) Symbiotic ammonium
transporter, partial (10%)
Length = 799
Score = 61.2 bits (147), Expect = 2e-10
Identities = 30/41 (73%), Positives = 35/41 (85%)
Frame = +3
Query: 124 KNLRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKVS 164
K +R+S + DHIMAER+RRQ LTERFIALSATIPGL KV+
Sbjct: 48 KKIRTSSQTIDHIMAERRRRQELTERFIALSATIPGLNKVN 170
>AW459618
Length = 372
Score = 57.8 bits (138), Expect = 3e-09
Identities = 29/44 (65%), Positives = 33/44 (74%)
Frame = +2
Query: 124 KNLRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKVSFLY 167
K +R SCE + HI+AERKRRQ LT IALSATIPGLKK+ Y
Sbjct: 17 KKVRRSCETRHHIIAERKRRQELTGSIIALSATIPGLKKMDKAY 148
>TC221759 homologue to UP|O81348 (O81348) Symbiotic ammonium transporter,
partial (70%)
Length = 818
Score = 52.8 bits (125), Expect = 8e-08
Identities = 24/43 (55%), Positives = 35/43 (80%)
Frame = +1
Query: 121 KRTKNLRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
KR K++ + ++ HI+AERKRR+ L++RFIALSA +PGLKK+
Sbjct: 562 KRLKHVLNCLNLKTHIIAERKRREKLSQRFIALSALVPGLKKM 690
>TC218502
Length = 873
Score = 52.8 bits (125), Expect = 8e-08
Identities = 25/43 (58%), Positives = 32/43 (74%)
Frame = +3
Query: 121 KRTKNLRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
K + RS QDHIMAERKRR+ L++ FIAL+A +PGLKK+
Sbjct: 30 KTSHGKRSPAHAQDHIMAERKRREKLSQSFIALAALVPGLKKM 158
>TC229276 weakly similar to UP|SRE2_HUMAN (Q12772) Sterol regulatory element
binding protein-2 (SREBP-2) (Sterol regulatory
element-binding transcription factor 2), partial (3%)
Length = 1069
Score = 50.8 bits (120), Expect = 3e-07
Identities = 38/92 (41%), Positives = 51/92 (55%), Gaps = 1/92 (1%)
Frame = +2
Query: 73 PSQYILSFENSTMQP-SPNSDIATCSSIMVQETTTLNNNVSSELPKIIKKRTKNLRSSCE 131
P ILSFE+ST+ P +P TC E ++ + E P +K K R +
Sbjct: 251 PKSCILSFEDSTLVPINPKK---TCQIYDHGE----HSKETQEKPHN-RKPLKRGRRFSQ 406
Query: 132 MQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
DHI+AERKRR+ ++ FIALSA IP LKK+
Sbjct: 407 TLDHILAERKRRENISRMFIALSALIPDLKKM 502
>TC233105
Length = 586
Score = 49.3 bits (116), Expect = 9e-07
Identities = 24/28 (85%), Positives = 25/28 (88%)
Frame = +3
Query: 135 HIMAERKRRQVLTERFIALSATIPGLKK 162
HIMAERKRRQ LT+ FIALSATIPGL K
Sbjct: 45 HIMAERKRRQQLTQSFIALSATIPGLNK 128
>CO985748
Length = 787
Score = 49.3 bits (116), Expect = 9e-07
Identities = 23/29 (79%), Positives = 26/29 (89%)
Frame = -1
Query: 133 QDHIMAERKRRQVLTERFIALSATIPGLK 161
+DHIM ERKRRQ++ ERFIALSA IPGLK
Sbjct: 355 RDHIMFERKRRQLMAERFIALSAIIPGLK 269
>AW394971
Length = 301
Score = 47.8 bits (112), Expect = 3e-06
Identities = 22/33 (66%), Positives = 27/33 (81%)
Frame = +2
Query: 131 EMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
E DHIM+ER +RQ LT +FIAL+A IPGLKK+
Sbjct: 2 EALDHIMSERNKRQELTSKFIALAAAIPGLKKM 100
>TC213205
Length = 590
Score = 39.7 bits (91), Expect = 7e-04
Identities = 21/55 (38%), Positives = 29/55 (52%)
Frame = +3
Query: 120 KKRTKNLRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKVSFLYKPTISLI 174
+K + RSS ++QDHIM ERKRR+ LT FI G +++ LI
Sbjct: 417 RKYKRGTRSSSQIQDHIMNERKRRENLTNMFIDFQPLFQGXXXSVYMFFSLFVLI 581
>TC213788 similar to GB|AAN73298.1|25141207|BT002301 At4g16430/dl4240w
{Arabidopsis thaliana;} , partial (19%)
Length = 452
Score = 37.0 bits (84), Expect = 0.005
Identities = 18/56 (32%), Positives = 33/56 (58%)
Frame = +1
Query: 108 NNNVSSELPKIIKKRTKNLRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
++++ ++ K K+ K E +H+ AER+RR+ L +RF AL A +P + K+
Sbjct: 244 SSSIHADERKPKKRGRKPANGREEPLNHVEAERQRREKLNQRFYALRAVVPNISKM 411
>TC209299 similar to GB|AAN73298.1|25141207|BT002301 At4g16430/dl4240w
{Arabidopsis thaliana;} , partial (31%)
Length = 608
Score = 37.0 bits (84), Expect = 0.005
Identities = 18/56 (32%), Positives = 33/56 (58%)
Frame = +3
Query: 108 NNNVSSELPKIIKKRTKNLRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
++++ ++ K K+ K E +H+ AER+RR+ L +RF AL A +P + K+
Sbjct: 393 SSSIHADERKPRKRGRKPANGREEPLNHVEAERQRREKLNQRFYALRAVVPNISKM 560
>TC213016 homologue to GB|AAN73298.1|25141207|BT002301 At4g16430/dl4240w
{Arabidopsis thaliana;} , partial (16%)
Length = 424
Score = 36.2 bits (82), Expect = 0.008
Identities = 17/44 (38%), Positives = 26/44 (58%)
Frame = +1
Query: 120 KKRTKNLRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
K+ K E +H+ AER+RR+ L +RF AL A +P + K+
Sbjct: 7 KRGRKPANGREEPLNHVEAERQRREKLNQRFYALRAVVPNISKM 138
>TC209106 similar to UP|Q41101 (Q41101) Phaseolin G-box binding protein PG1,
partial (40%)
Length = 1101
Score = 35.8 bits (81), Expect = 0.011
Identities = 17/44 (38%), Positives = 26/44 (58%)
Frame = +3
Query: 120 KKRTKNLRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
K+ K E +H+ AER+RR+ L +RF AL A +P + K+
Sbjct: 267 KRGRKPANGREEPLNHVEAERQRREKLNQRFYALRAVVPNVSKM 398
>TC218174 similar to UP|Q41102 (Q41102) Phaseolin G-box binding protein PG2
(Fragment), partial (49%)
Length = 1345
Score = 35.4 bits (80), Expect = 0.014
Identities = 15/33 (45%), Positives = 23/33 (69%)
Frame = +3
Query: 131 EMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
E +H+ AER+RR+ L +RF AL A +P + K+
Sbjct: 423 EPLNHVEAERQRREKLNQRFYALRAVVPNVSKM 521
>TC218173 similar to UP|Q41102 (Q41102) Phaseolin G-box binding protein PG2
(Fragment), partial (28%)
Length = 867
Score = 35.4 bits (80), Expect = 0.014
Identities = 15/33 (45%), Positives = 23/33 (69%)
Frame = +3
Query: 131 EMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
E +H+ AER+RR+ L +RF AL A +P + K+
Sbjct: 21 EPLNHVEAERQRREKLNQRFYALRAVVPNVSKM 119
>TC213483 similar to UP|Q8LPT0 (Q8LPT0) At2g46510/F13A10.4, partial (17%)
Length = 914
Score = 35.0 bits (79), Expect = 0.018
Identities = 16/44 (36%), Positives = 26/44 (58%)
Frame = +2
Query: 120 KKRTKNLRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
K+ K E +H+ AER+RR+ L +RF AL + +P + K+
Sbjct: 113 KRGRKPANGREEPLNHVEAERQRREKLNQRFYALRSVVPNISKM 244
>TC230761 weakly similar to UP|O81348 (O81348) Symbiotic ammonium
transporter, partial (13%)
Length = 697
Score = 34.3 bits (77), Expect = 0.031
Identities = 15/23 (65%), Positives = 19/23 (82%)
Frame = +1
Query: 140 RKRRQVLTERFIALSATIPGLKK 162
+KR++ L ERF+ALSATIPG K
Sbjct: 106 KKRKRELAERFLALSATIPGFTK 174
>TC231689 weakly similar to UP|Q38736 (Q38736) DEL, partial (22%)
Length = 937
Score = 33.9 bits (76), Expect = 0.041
Identities = 12/29 (41%), Positives = 21/29 (72%)
Frame = +2
Query: 134 DHIMAERKRRQVLTERFIALSATIPGLKK 162
+H+M+ER+RR L +RF+ L + +P + K
Sbjct: 707 NHVMSERRRRAKLNQRFLTLRSMVPSISK 793
>TC215736 similar to GB|AAO63377.1|28950907|BT005313 At4g34530 {Arabidopsis
thaliana;} , partial (28%)
Length = 1047
Score = 33.9 bits (76), Expect = 0.041
Identities = 26/83 (31%), Positives = 41/83 (49%), Gaps = 9/83 (10%)
Frame = +1
Query: 91 SDIATCSSIMV----QETTT-----LNNNVSSELPKIIKKRTKNLRSSCEMQDHIMAERK 141
S I C++I +ET T N+ +SE P I R + +++ H +AER
Sbjct: 196 SKITKCNTINTNTNNKETCTDTSNSKQNSKASEKPDYIHVRARRGQAT---DSHSLAERV 366
Query: 142 RRQVLTERFIALSATIPGLKKVS 164
RR+ ++ER L +PG KV+
Sbjct: 367 RREKISERMNYLQDLVPGCNKVT 435
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.127 0.354
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,246,888
Number of Sequences: 63676
Number of extensions: 90190
Number of successful extensions: 581
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 578
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 580
length of query: 176
length of database: 12,639,632
effective HSP length: 91
effective length of query: 85
effective length of database: 6,845,116
effective search space: 581834860
effective search space used: 581834860
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC124966.6


