

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124966.14 - phase: 0
(588 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
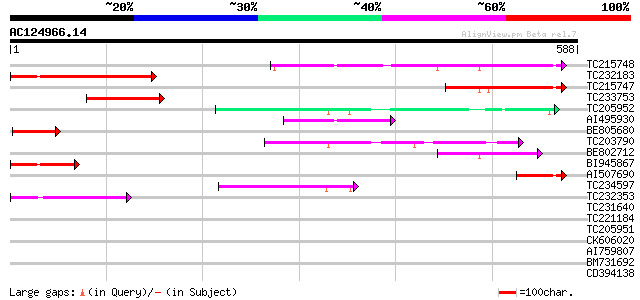
Score E
Sequences producing significant alignments: (bits) Value
TC215748 similar to GB|AAP37658.1|30725272|BT008299 At2g25430/F1... 219 3e-57
TC232183 142 5e-34
TC215747 similar to GB|AAP37658.1|30725272|BT008299 At2g25430/F1... 111 1e-24
TC233753 similar to GB|AAP37658.1|30725272|BT008299 At2g25430/F1... 106 3e-23
TC205952 103 2e-22
AI495930 87 3e-17
BE805680 77 3e-14
TC203790 76 5e-14
BE802712 weakly similar to GP|18700099|gb| AT4g32280/F10M6_80 {A... 62 9e-10
BI945867 61 1e-09
AI507690 59 5e-09
TC234597 weakly similar to UP|Q7ZRD4 (Q7ZRD4) Envelope glycoprot... 57 2e-08
TC232353 similar to UP|Q7X9S1 (Q7X9S1) Fiber protein Fb19 (Fragm... 48 1e-05
TC231640 homologue to UP|Q918P0 (Q918P0) Latency associated anti... 42 0.001
TC221184 similar to UP|O22179 (O22179) MYB family transcription ... 40 0.002
TC205951 weakly similar to GB|AAD12692.3|20197544|AC006069 expre... 40 0.002
CK606020 40 0.003
AI759807 39 0.006
BM731692 37 0.019
CD394138 37 0.019
>TC215748 similar to GB|AAP37658.1|30725272|BT008299 At2g25430/F13B15.9
{Arabidopsis thaliana;} , partial (47%)
Length = 1413
Score = 219 bits (557), Expect = 3e-57
Identities = 138/332 (41%), Positives = 192/332 (57%), Gaps = 25/332 (7%)
Frame = +2
Query: 271 PEY---YKVYDVFCRVGKQYDELDLFYSWSKSIGIGRSSEYPEIEKVTTKKLDLMDQFIR 327
PEY K +D + KQ DEL FY+W K G+ RSSEYPE++K+T+K L+ +++F+R
Sbjct: 2 PEYADCVKAFDAYASAAKQIDELVDFYNWCKDTGVARSSEYPEVQKITSKLLETLEEFVR 181
Query: 328 DKSLVSQANKLITQEENNEKNEEENEVEEDMNEIKALPPPEGFNEEQVEE-EIKEQDQKE 386
D++ ++ + +EE + E E DMNEIKALPPPE + E E K Q Q
Sbjct: 182 DRAKRPKSPE--RKEEAPPVEKVEEEPAPDMNEIKALPPPENYTPPPPPEPEPKPQPQVT 355
Query: 387 EEKIVQTEGDLLDLTDSMTNQDYVGNKLALALFDELP-NTTSNTIQALPWHAFDDVS--- 442
E DL++L D D GNK ALALF P N + + +A P + +V+
Sbjct: 356 E--------DLVNLRDDAVTADDQGNKFALALFAGAPANNANGSWEAFPSNGQPEVTSAW 511
Query: 443 ----------DWETTLVQSSTNLPNQKPSLGGGFDTLLLDSMYNQKPSLQGMN----GYG 488
DWE LV++++NL QK +LGGG D LLL MY+Q Q ++ G
Sbjct: 512 QTPAAEPGKADWELALVETASNLSKQKAALGGGLDPLLLTGMYDQGMVRQHVSTTQLSGG 691
Query: 489 SASSVAIRS--EATMLALPAPPTS-RNGSQDPFAASMLVAPPAYVQMSEMETRQRLLAEE 545
SASSVA+ + +LALPAP S + +QDPFAAS+ V PP+YVQM++ME +Q LL +E
Sbjct: 692 SASSVALPGPGKTPVLALPAPDGSVQPVNQDPFAASLSVPPPSYVQMADMEKKQHLLVQE 871
Query: 546 QAIWQQYAKNGMQGQVGFATQQQPNSNFYMGG 577
Q +W QYA++GMQGQ A + + +Y GG
Sbjct: 872 QQVWHQYARDGMQGQSSLA--KLNGAGYYAGG 961
>TC232183
Length = 598
Score = 142 bits (357), Expect = 5e-34
Identities = 71/153 (46%), Positives = 108/153 (70%), Gaps = 2/153 (1%)
Frame = +3
Query: 2 SQSTLRRAIGAVKDQTSIGIAKVGSSASIGDLQVAIVKATKHDENPAEERHIREILSLTC 61
+Q +LR+A+GA+KD T++ +AKV S +L +AIV+AT H E PA+E+HIR I S
Sbjct: 141 TQKSLRKALGALKDTTTVSLAKVNSDYK--ELDIAIVRATNHVERPAKEKHIRAIFSAIS 314
Query: 62 YSR--AFISSCVNTLSKRLIKTSSWTVALKTLVLIQRLLADGDRAYEQEIFFSTQRGTRL 119
+R A ++ C++ L++RL KT +W VALKTL++I R L + D + +E+ + + +
Sbjct: 315 ATRPRADVAYCIHALARRLSKTHNWAVALKTLIVIHRALREVDPTFHEELINYGRSRSHM 494
Query: 120 LNMSDFRDKSKSNSWDYSSFVRTYALYLDERLE 152
LNM+ F+D S N+WDYS++VRTYAL+L+ERLE
Sbjct: 495 LNMAHFKDDSSPNAWDYSAWVRTYALFLEERLE 593
>TC215747 similar to GB|AAP37658.1|30725272|BT008299 At2g25430/F13B15.9
{Arabidopsis thaliana;} , partial (21%)
Length = 799
Score = 111 bits (277), Expect = 1e-24
Identities = 65/134 (48%), Positives = 88/134 (65%), Gaps = 9/134 (6%)
Frame = +3
Query: 453 TNLPNQKPSLGGGFDTLLLDSMYNQKPSLQGMN----GYGSASSVAI----RSEATMLAL 504
+NL QK +LGGGFD LLL MY+Q Q ++ GSASSVA+ ++ +LAL
Sbjct: 3 SNLSKQKATLGGGFDPLLLTGMYDQGMVRQHVSTTQLSGGSASSVALPGPGKTTTPVLAL 182
Query: 505 PAPPTS-RNGSQDPFAASMLVAPPAYVQMSEMETRQRLLAEEQAIWQQYAKNGMQGQVGF 563
PAP S + +QDPFAAS+ V PP+YVQM++ME +Q LL +EQ +W QYA++GMQGQ
Sbjct: 183 PAPDGSVQPVNQDPFAASLSVPPPSYVQMADMEKKQHLLVQEQQVWHQYARDGMQGQSSL 362
Query: 564 ATQQQPNSNFYMGG 577
A + + +Y GG
Sbjct: 363 A--KLNGAGYYAGG 398
>TC233753 similar to GB|AAP37658.1|30725272|BT008299 At2g25430/F13B15.9
{Arabidopsis thaliana;} , partial (14%)
Length = 429
Score = 106 bits (264), Expect = 3e-23
Identities = 48/81 (59%), Positives = 66/81 (81%)
Frame = +2
Query: 80 KTSSWTVALKTLVLIQRLLADGDRAYEQEIFFSTQRGTRLLNMSDFRDKSKSNSWDYSSF 139
KT W VALK L+L+ RL+ DG +++EI ++T+RGTRLLNMSDFRD++ S+SWD+S+F
Sbjct: 8 KTRDWIVALKALMLVHRLMNDGPPLFQEEILYATRRGTRLLNMSDFRDEAHSSSWDHSAF 187
Query: 140 VRTYALYLDERLEYRMQYKRG 160
VRTYALYLD+RLE + ++G
Sbjct: 188 VRTYALYLDQRLELMLFDRKG 250
>TC205952
Length = 1414
Score = 103 bits (258), Expect = 2e-22
Identities = 100/393 (25%), Positives = 158/393 (39%), Gaps = 36/393 (9%)
Frame = +1
Query: 214 LQLLLERFMACRPTGRAKTHRMVIVALYPIVKESFQTYHDMTSILGIFIDRFTEMEVPEY 273
LQ LL R + CRP G A ++ ++ AL ++KESF+ Y + + +D+F EM E
Sbjct: 10 LQQLLYRLVGCRPEGAAVSNYVIQYALALVLKESFKIYCAINDGIINLVDKFFEMPRHEA 189
Query: 274 YKVYDVFCRVGKQYDELDLFYSWSKSIGIGRSSEYPEIEKVTTKKLDLMDQFIRDK---- 329
K + + R G+Q L FY K + + R+ ++P + + L M+++I++
Sbjct: 190 IKALEAYKRAGQQAASLSDFYDVCKGLELARNFQFPVLREPPQSFLTTMEEYIKEAPRVV 369
Query: 330 --------SLVSQANKLITQEENNEKNEEE--------NEVEEDMNEIKALPPPEGFNEE 373
L + +++ E+ +EE+ N V D PPP N
Sbjct: 370 TVPTEPLLQLTYRPEEVLAIEDAKPSDEEQEPPVPVDNNVVVSDSEPAPPPPPPSAHNNF 549
Query: 374 QVEEEIKEQDQKEEEKIVQTEGDLLDLTDSMTNQDYV--GNKLALALFDELPNTTSNTIQ 431
+ GDLL L D+ + + N LALA+ TTS
Sbjct: 550 ET-------------------GDLLGLNDTAPDASSIEERNALALAIVPTETGTTSAFNT 672
Query: 432 ALPWHAFDDVSDWETTLVQS-STNL-PNQKPSLGGGFDTLLLDSMYNQKPSLQGMNGYGS 489
D + WE LV + ST++ + L GG D+L L+S+Y++
Sbjct: 673 TAAQTKDFDPTGWELALVSTPSTDISAANERQLAGGLDSLTLNSLYDE------------ 816
Query: 490 ASSVAIRSEATMLALPAPPTSRNGSQDPFAASMLVAPPAYVQMSEMETRQRLLAEEQAIW 549
A RS+ + PAP QDPFA S + PP VQ++ M+ + Q +
Sbjct: 817 ---AAYRSQQPVYGAPAPNPFE--MQDPFALSSSIPPPPAVQLAAMQQQANPFGPYQQPF 981
Query: 550 QQYAKNGMQ------------GQVGFATQQQPN 570
Q + Q G GF PN
Sbjct: 982 QPQPQPQQQQHHMLMNPANPFGDAGFGAFPAPN 1080
>AI495930
Length = 357
Score = 86.7 bits (213), Expect = 3e-17
Identities = 47/117 (40%), Positives = 70/117 (59%), Gaps = 1/117 (0%)
Frame = +3
Query: 285 KQYDELDLFYSWSKSIGIGRSSEYPEIEKVTTKKLDLMDQFIRDKSLVSQANKLITQEEN 344
KQ DEL FY+W K G+ RSSEYPE++++T K L+ +++F+RD++ ++ + +EE
Sbjct: 9 KQIDELVAFYNWCKDTGVARSSEYPEVQRITNKLLETLEEFVRDRAKRPKSPE--RKEEV 182
Query: 345 NEKNEEENEVEEDMNEIKALPPPEGF-NEEQVEEEIKEQDQKEEEKIVQTEGDLLDL 400
+ E E DMNEIKALPPPE + E E K Q Q E+ + + LL +
Sbjct: 183 PPVEKVEEEPAPDMNEIKALPPPENYIPPPPPEPEPKPQPQVTEDLVNLRDDQLLQM 353
>BE805680
Length = 417
Score = 76.6 bits (187), Expect = 3e-14
Identities = 37/49 (75%), Positives = 42/49 (85%)
Frame = +3
Query: 4 STLRRAIGAVKDQTSIGIAKVGSSASIGDLQVAIVKATKHDENPAEERH 52
S RRA+GAVKDQTSI +AKVGSS S+ DL VAIVKAT+HDE PAEE+H
Sbjct: 270 SKFRRALGAVKDQTSISLAKVGSSTSLADLDVAIVKATRHDEYPAEEKH 416
>TC203790
Length = 867
Score = 75.9 bits (185), Expect = 5e-14
Identities = 75/280 (26%), Positives = 116/280 (40%), Gaps = 11/280 (3%)
Frame = +1
Query: 265 FTEMEVPEYYKVYDVFCRVGKQYDELDLFYSWSKSIGIGRSSEYPEIEKVTTKKLDLMDQ 324
F EM+ + K D++ RVG Q + L FY +++ IGR ++ ++E+ + L M++
Sbjct: 106 FFEMQRHDALKALDIYRRVGHQAERLSEFYEICRNLDIGRGEKFIKVEQPPSSFLQAMEE 285
Query: 325 FIRDK---SLVSQANKLITQEENNEKNEEENEVEEDMNEIKALPPPEGFNEEQVEEEIKE 381
+++D +V + + +E + ++ EVEE+ + PP E V
Sbjct: 286 YVKDAPQGPIVRKDQAIENKEVLAIEYKKTTEVEEECPPSPSPSPPPPPPSEPV------ 447
Query: 382 QDQKEEEKIVQTEGDLLDLTDSMTNQDYVGNKLALAL------FDELPNTTSNTIQALPW 435
K E VQ DLL+L D + + K ALAL ++ P+ SN
Sbjct: 448 ---KVEAPPVQPPPDLLNLEDPVPAAAELEEKNALALAIVPVAVEQQPSAASNQA----- 603
Query: 436 HAFDDVSDWETTLV--QSSTNLPNQKPSLGGGFDTLLLDSMYNQKPSLQGMNGYGSASSV 493
+ + WE LV SS L GG D L LDS+Y+ N +
Sbjct: 604 ---NGTTGWELALVTAPSSNETATAASKLAGGLDKLTLDSLYDDALRRNNQNVSYNPWEP 774
Query: 494 AIRSEATMLALPAPPTSRNGSQDPFAASMLVAPPAYVQMS 533
A P + DPF AS VA P VQM+
Sbjct: 775 A----------PGGNMMQPTMHDPFFASNTVAAPPSVQMA 864
>BE802712 weakly similar to GP|18700099|gb| AT4g32280/F10M6_80 {Arabidopsis
thaliana}, partial (7%)
Length = 367
Score = 61.6 bits (148), Expect = 9e-10
Identities = 40/116 (34%), Positives = 58/116 (49%), Gaps = 7/116 (6%)
Frame = +2
Query: 444 WETTLVQSSTNLPNQKPSLGGGFDTLLLDSMYNQKPSLQGMN------GYGSASSVAIRS 497
WE T ++ L + ++GGG D LLL M++Q Q ++ G ++ +
Sbjct: 20 WELT*GDTAITLSDHNAAVGGGLDPLLLTDMFDQGMVKQHVSTTLLSGGSACNDALPVPG 199
Query: 498 EATMLALPAPPTS-RNGSQDPFAASMLVAPPAYVQMSEMETRQRLLAEEQAIWQQY 552
+ T L L A S + +QDPF AS+ V PP+Y QM+ ME QR L E W Y
Sbjct: 200 KTTDLGLTALDGSVQTDNQDPFDASVSVLPPSYGQMANMENTQR*LVHEMHGWYNY 367
>BI945867
Length = 411
Score = 61.2 bits (147), Expect = 1e-09
Identities = 32/73 (43%), Positives = 50/73 (67%), Gaps = 2/73 (2%)
Frame = +1
Query: 2 SQSTLRRAIGAVKDQTSIGIAKVGSSASIGDLQVAIVKATKHDENPAEERHIREILSLTC 61
+Q +LR+A+GA+KD T++ +AKV S +L +AIV+AT H E PA+E+HIR I S
Sbjct: 196 TQKSLRKALGALKDTTTVSLAKVNSDYK--ELDIAIVRATNHVERPAKEKHIRAIFSAIS 369
Query: 62 YS--RAFISSCVN 72
+ RA ++ C++
Sbjct: 370 ATRPRADVAYCIH 408
>AI507690
Length = 429
Score = 59.3 bits (142), Expect = 5e-09
Identities = 26/52 (50%), Positives = 37/52 (71%)
Frame = +1
Query: 526 PPAYVQMSEMETRQRLLAEEQAIWQQYAKNGMQGQVGFATQQQPNSNFYMGG 577
PP+YVQM++ME +Q LL +EQ +W QYA++GMQGQ A + + +Y GG
Sbjct: 4 PPSYVQMADMEKKQHLLVQEQQVWHQYARDGMQGQSSLA--KLNGAGYYAGG 153
>TC234597 weakly similar to UP|Q7ZRD4 (Q7ZRD4) Envelope glycoprotein
(Fragment), partial (5%)
Length = 822
Score = 57.0 bits (136), Expect = 2e-08
Identities = 42/173 (24%), Positives = 73/173 (41%), Gaps = 28/173 (16%)
Frame = +3
Query: 217 LLERFMACRPTGRAKTHRMVIVALYPIVKESFQTYHDMTSILGIFIDRFTEMEVPEYYKV 276
L++R M C P G A +V VA+ I+++SF Y + +D EM
Sbjct: 6 LIDRVMECYPVGVAARSFIVQVAMKLIIRDSFVCYTKFRREIVTVLDNLLEMPYRNCIAA 185
Query: 277 YDVFCRVGKQYDELDLFYSWSKSIGIGRSSEYPEIEKVTTKKLDLMDQFIR--------- 327
++++ + Q +EL FY W K+ G+ EYP +E + ++ ++ F+
Sbjct: 186 FNIYKKAAAQTNELYEFYEWCKAKGLCGMYEYPLVEPIPYIQIKALESFLSGMWQLTESS 365
Query: 328 --------------DKSLVSQANKLITQEENNEKNEEEN-----EVEEDMNEI 361
S SQ ++ ++ N K EEE E+EED ++
Sbjct: 366 TSPTTSTSPSSSGDSPSNFSQGQIVMRRDVVNIKGEEEKPLIELELEEDNGDV 524
>TC232353 similar to UP|Q7X9S1 (Q7X9S1) Fiber protein Fb19 (Fragment),
partial (33%)
Length = 725
Score = 47.8 bits (112), Expect = 1e-05
Identities = 35/127 (27%), Positives = 63/127 (49%), Gaps = 2/127 (1%)
Frame = +2
Query: 2 SQSTLRRAIGAVKDQTSIGIAKVGSSASIGDLQVAIVKATKHDEN-PAEERHIREILSLT 60
+ + L IG +KD+ S A + S + + +++AT HD + P +H+ +LS
Sbjct: 356 NMTKLTHLIGIIKDKASQSKAALLSKRTT----LFLLRATSHDSSTPPTRKHLATLLSSG 523
Query: 61 CYSRAFISSCVNTLSKRLIKTSSWTVALKTLVLIQRLLADGDRAYEQEI-FFSTQRGTRL 119
SRA S+ V L RL T++ VALK L+ + ++ + ++ + +
Sbjct: 524 DGSRATASAAVEVLMDRLQGTNNAAVALKCLIAVHHIIHHXSFILQDQLXVYPSAXXRNY 703
Query: 120 LNMSDFR 126
LN+S+FR
Sbjct: 704 LNLSNFR 724
>TC231640 homologue to UP|Q918P0 (Q918P0) Latency associated antigen, partial
(7%)
Length = 466
Score = 41.6 bits (96), Expect = 0.001
Identities = 26/63 (41%), Positives = 36/63 (56%)
Frame = +1
Query: 340 TQEENNEKNEEENEVEEDMNEIKALPPPEGFNEEQVEEEIKEQDQKEEEKIVQTEGDLLD 399
TQEE EK EEE E EE EE+ EEE +E+++KEEE+ + EG++L
Sbjct: 19 TQEEEEEKEEEEEEEEE---------------EEEEEEEEEEEEEKEEEE--EEEGNVLV 147
Query: 400 LTD 402
+ D
Sbjct: 148 IID 156
>TC221184 similar to UP|O22179 (O22179) MYB family transcription factor
(At2g23280) (MYB transcription factor), partial (15%)
Length = 705
Score = 40.4 bits (93), Expect = 0.002
Identities = 22/79 (27%), Positives = 44/79 (54%)
Frame = +3
Query: 346 EKNEEENEVEEDMNEIKALPPPEGFNEEQVEEEIKEQDQKEEEKIVQTEGDLLDLTDSMT 405
++NEEE + + PP E EE+ EEE +E+++KE+E++ Q +++ +T
Sbjct: 228 KENEEERVLSPVTTSLSLFPPGEKSEEEEEEEE-QEEEEKEKEELFQVN---VNVNQKVT 395
Query: 406 NQDYVGNKLALALFDELPN 424
+Q+Y + + +E+ N
Sbjct: 396 DQNYFMQMMQRMIAEEVRN 452
>TC205951 weakly similar to GB|AAD12692.3|20197544|AC006069 expressed protein
{Arabidopsis thaliana;} , partial (22%)
Length = 782
Score = 40.4 bits (93), Expect = 0.002
Identities = 38/141 (26%), Positives = 59/141 (40%), Gaps = 2/141 (1%)
Frame = +1
Query: 440 DVSDWETTLVQS-STNLPN-QKPSLGGGFDTLLLDSMYNQKPSLQGMNGYGSASSVAIRS 497
D + WE LV + ST++ + L GG +L L+S+Y++ Y SA
Sbjct: 16 DPTGWELALVSTPSTDISAANERQLAGGLHSLTLNSLYDEA-------AYRSA------- 153
Query: 498 EATMLALPAPPTSRNGSQDPFAASMLVAPPAYVQMSEMETRQRLLAEEQAIWQQYAKNGM 557
+ + PAP QDPFA S + PP VQM+ M+ + Q +Q +
Sbjct: 154 QQPVYGAPAPNPFE--VQDPFALSSSIPPPPAVQMAAMQQQANPFGSYQQPFQPQPQLQQ 327
Query: 558 QGQVGFATQQQPNSNFYMGGY 578
Q Q+ P + G +
Sbjct: 328 QQQLMLMNPANPFGDAGFGAF 390
>CK606020
Length = 340
Score = 40.0 bits (92), Expect = 0.003
Identities = 21/50 (42%), Positives = 32/50 (64%)
Frame = +1
Query: 339 ITQEENNEKNEEENEVEEDMNEIKALPPPEGFNEEQVEEEIKEQDQKEEE 388
+ +EE E+ EEE E EE+ EI+ E EE+VEEE++E++ + EE
Sbjct: 193 LDEEEEEEEEEEEEEEEEEEEEIEEEEEEE---EEEVEEEVEEEEVEGEE 333
Score = 31.6 bits (70), Expect = 1.0
Identities = 17/49 (34%), Positives = 30/49 (60%)
Frame = +1
Query: 342 EENNEKNEEENEVEEDMNEIKALPPPEGFNEEQVEEEIKEQDQKEEEKI 390
+E + +EE E EE+ E + EE+ EEEI+E++++EEE++
Sbjct: 175 DEIDPSLDEEEEEEEEEEEEE---------EEEEEEEIEEEEEEEEEEV 294
Score = 30.0 bits (66), Expect = 3.0
Identities = 18/47 (38%), Positives = 27/47 (57%)
Frame = +1
Query: 348 NEEENEVEEDMNEIKALPPPEGFNEEQVEEEIKEQDQKEEEKIVQTE 394
+EEE E EE+ E EE+ EEE E++++EEE+ V+ E
Sbjct: 196 DEEEEEEEEEEEE-----------EEEEEEEEIEEEEEEEEEEVEEE 303
>AI759807
Length = 207
Score = 38.9 bits (89), Expect = 0.006
Identities = 17/34 (50%), Positives = 24/34 (70%)
Frame = +1
Query: 169 EDEEEQSRESKRERYRERDRDKEIVVRSTPLREM 202
E E E+ RE +RER RER+R++E+V S P R +
Sbjct: 22 ERERERERERERERERERERERELVPNSAPCRHL 123
Score = 31.6 bits (70), Expect = 1.0
Identities = 13/24 (54%), Positives = 19/24 (79%)
Frame = +3
Query: 168 DEDEEEQSRESKRERYRERDRDKE 191
+E E E+ RE +RER RER+R++E
Sbjct: 3 EERERERERERERERERERERERE 74
Score = 30.8 bits (68), Expect = 1.8
Identities = 13/23 (56%), Positives = 18/23 (77%)
Frame = +3
Query: 169 EDEEEQSRESKRERYRERDRDKE 191
E E E+ RE +RER RER+R++E
Sbjct: 12 ERERERERERERERERERERERE 80
Score = 30.8 bits (68), Expect = 1.8
Identities = 13/23 (56%), Positives = 18/23 (77%)
Frame = +3
Query: 169 EDEEEQSRESKRERYRERDRDKE 191
E E E+ RE +RER RER+R++E
Sbjct: 18 ERERERERERERERERERERERE 86
Score = 30.8 bits (68), Expect = 1.8
Identities = 13/23 (56%), Positives = 18/23 (77%)
Frame = +2
Query: 169 EDEEEQSRESKRERYRERDRDKE 191
E E E+ RE +RER RER+R++E
Sbjct: 20 ERERERERERERERERERERERE 88
Score = 30.8 bits (68), Expect = 1.8
Identities = 13/23 (56%), Positives = 18/23 (77%)
Frame = +2
Query: 169 EDEEEQSRESKRERYRERDRDKE 191
E E E+ RE +RER RER+R++E
Sbjct: 14 ERERERERERERERERERERERE 82
Score = 30.8 bits (68), Expect = 1.8
Identities = 13/23 (56%), Positives = 18/23 (77%)
Frame = +2
Query: 169 EDEEEQSRESKRERYRERDRDKE 191
E E E+ RE +RER RER+R++E
Sbjct: 8 ERERERERERERERERERERERE 76
>BM731692
Length = 293
Score = 37.4 bits (85), Expect = 0.019
Identities = 19/37 (51%), Positives = 27/37 (72%), Gaps = 2/37 (5%)
Frame = +2
Query: 169 EDEEEQSRESKRERYRERDRDKEIVVR--STPLREMK 203
E E E+ RE +RER RER+R++EI + +TPL +MK
Sbjct: 29 EREREREREREREREREREREREIYTKRQATPLWKMK 139
Score = 32.7 bits (73), Expect = 0.46
Identities = 15/28 (53%), Positives = 20/28 (70%)
Frame = +3
Query: 169 EDEEEQSRESKRERYRERDRDKEIVVRS 196
E E E+ RE +RER RER+R++E RS
Sbjct: 27 ERERERERERERERERERERERERFTRS 110
Score = 31.6 bits (70), Expect = 1.0
Identities = 14/30 (46%), Positives = 20/30 (66%)
Frame = +1
Query: 169 EDEEEQSRESKRERYRERDRDKEIVVRSTP 198
E E E+ RE +RER RER+R++E + P
Sbjct: 25 ERERERERERERERERERERERERDLHEAP 114
Score = 31.6 bits (70), Expect = 1.0
Identities = 17/51 (33%), Positives = 27/51 (52%)
Frame = +1
Query: 169 EDEEEQSRESKRERYRERDRDKEIVVRSTPLREMKTDDLFSRMQHLQLLLE 219
E E E+ RE +RER RER+RD T L ++ S + ++++E
Sbjct: 37 ERERERERERERERERERERDLHEAPGDTTLENEMKAEVQSDAKSREIIIE 189
Score = 30.8 bits (68), Expect = 1.8
Identities = 13/23 (56%), Positives = 18/23 (77%)
Frame = +3
Query: 169 EDEEEQSRESKRERYRERDRDKE 191
E E E+ RE +RER RER+R++E
Sbjct: 9 ERERERERERERERERERERERE 77
Score = 30.8 bits (68), Expect = 1.8
Identities = 13/23 (56%), Positives = 18/23 (77%)
Frame = +3
Query: 169 EDEEEQSRESKRERYRERDRDKE 191
E E E+ RE +RER RER+R++E
Sbjct: 21 ERERERERERERERERERERERE 89
Score = 30.8 bits (68), Expect = 1.8
Identities = 13/23 (56%), Positives = 18/23 (77%)
Frame = +3
Query: 169 EDEEEQSRESKRERYRERDRDKE 191
E E E+ RE +RER RER+R++E
Sbjct: 15 ERERERERERERERERERERERE 83
Score = 30.8 bits (68), Expect = 1.8
Identities = 13/23 (56%), Positives = 18/23 (77%)
Frame = +2
Query: 169 EDEEEQSRESKRERYRERDRDKE 191
E E E+ RE +RER RER+R++E
Sbjct: 5 ERERERERERERERERERERERE 73
Score = 30.8 bits (68), Expect = 1.8
Identities = 13/23 (56%), Positives = 18/23 (77%)
Frame = +1
Query: 169 EDEEEQSRESKRERYRERDRDKE 191
E E E+ RE +RER RER+R++E
Sbjct: 1 ERERERERERERERERERERERE 69
>CD394138
Length = 611
Score = 37.4 bits (85), Expect = 0.019
Identities = 19/44 (43%), Positives = 29/44 (65%), Gaps = 3/44 (6%)
Frame = +2
Query: 151 LEYRMQYKRGR---SGRFAYDEDEEEQSRESKRERYRERDRDKE 191
L Y + + RG+ S R ++E E E+ RE +RER RER+R++E
Sbjct: 419 LNYDLGHPRGQECASCRIRHEERERERERERERERERERERERE 550
Score = 32.7 bits (73), Expect = 0.46
Identities = 16/33 (48%), Positives = 21/33 (63%)
Frame = +1
Query: 159 RGRSGRFAYDEDEEEQSRESKRERYRERDRDKE 191
RG R E E E+ RE +RER RER+R++E
Sbjct: 478 RGERERERERERERERERERERERERERERERE 576
Score = 32.0 bits (71), Expect = 0.79
Identities = 17/44 (38%), Positives = 25/44 (56%)
Frame = +2
Query: 148 DERLEYRMQYKRGRSGRFAYDEDEEEQSRESKRERYRERDRDKE 191
+ER R + + R E E E+ RE +RER RER+R++E
Sbjct: 479 EERERERERERERERERERERERERERERERERERERERERERE 610
Score = 31.6 bits (70), Expect = 1.0
Identities = 18/42 (42%), Positives = 26/42 (61%)
Frame = +3
Query: 150 RLEYRMQYKRGRSGRFAYDEDEEEQSRESKRERYRERDRDKE 191
R E+ + +R R R E E E+ RE +RER RER+R++E
Sbjct: 462 RAEFGTRRERERE-RERERERERERERERERERERERERERE 584
Score = 31.2 bits (69), Expect = 1.3
Identities = 17/42 (40%), Positives = 24/42 (56%)
Frame = +2
Query: 150 RLEYRMQYKRGRSGRFAYDEDEEEQSRESKRERYRERDRDKE 191
R E R + + R E E E+ RE +RER RER+R++E
Sbjct: 473 RHEERERERERERERERERERERERERERERERERERERERE 598
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.131 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,901,879
Number of Sequences: 63676
Number of extensions: 274619
Number of successful extensions: 10914
Number of sequences better than 10.0: 311
Number of HSP's better than 10.0 without gapping: 2817
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6245
length of query: 588
length of database: 12,639,632
effective HSP length: 103
effective length of query: 485
effective length of database: 6,081,004
effective search space: 2949286940
effective search space used: 2949286940
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC124966.14


