

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
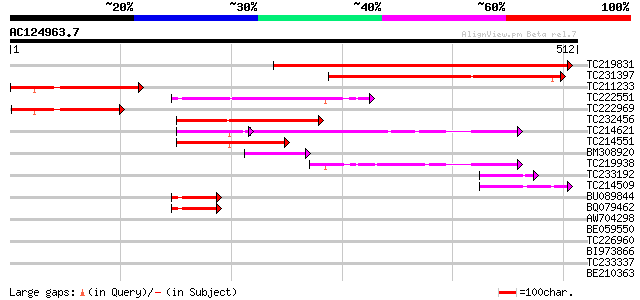
Query= AC124963.7 + phase: 0
(512 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC219831 weakly similar to GB|AAS55571.1|45237181|BT011759 At3g0... 460 e-130
TC231397 weakly similar to GB|AAP21157.1|30102478|BT006349 At5g0... 271 7e-73
TC211233 weakly similar to GB|AAP21157.1|30102478|BT006349 At5g0... 152 5e-37
TC222551 similar to GB|AAS49080.1|44917511|BT011717 At3g44520 {A... 117 1e-26
TC222969 114 1e-25
TC232456 112 4e-25
TC214621 similar to UP|Q84VY8 (Q84VY8) At2g36290, partial (62%) 84 8e-23
TC214551 similar to UP|Q9SJM9 (Q9SJM9) Expressed protein, partia... 81 1e-15
BM308920 56 3e-08
TC219938 53 3e-07
TC233192 weakly similar to UP|Q84VY8 (Q84VY8) At2g36290, partial... 46 3e-05
TC214509 similar to UP|Q84VY8 (Q84VY8) At2g36290, partial (31%) 42 5e-04
BU089844 similar to PIR|A86217|A862 protein T23G18.18 [imported]... 42 5e-04
BQ079462 similar to PIR|A86217|A862 protein T23G18.18 [imported]... 42 5e-04
AW704298 similar to PIR|A86217|A862 protein T23G18.18 [imported]... 41 0.001
BE059550 40 0.003
TC226960 similar to UP|Q9ZP87 (Q9ZP87) Epoxide hydrolase, partia... 39 0.004
BI973866 weakly similar to GP|1354849|gb| epoxide hydrolase {Nic... 39 0.007
TC233337 similar to GB|AAO63446.1|28951045|BT005382 At1g64670 {A... 37 0.028
BE210363 34 0.14
>TC219831 weakly similar to GB|AAS55571.1|45237181|BT011759 At3g09690
{Arabidopsis thaliana;} , partial (45%)
Length = 1032
Score = 460 bits (1183), Expect = e-130
Identities = 210/270 (77%), Positives = 242/270 (88%)
Frame = +1
Query: 239 LVDAVNVTDKFWVLCHSSGCIHAWASLKYIPERIAGAAMLAPMVSPYESHMTKDEMKRTW 298
LV+AVNVTDKFW+LCHSSGCIHAWASL+YIPE+IAGAAMLAPM++PY+ HMTK+EMKRTW
Sbjct: 1 LVNAVNVTDKFWILCHSSGCIHAWASLRYIPEKIAGAAMLAPMINPYDPHMTKEEMKRTW 180
Query: 299 EKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKHERIDKQFSLSLGKKDEILVDEPAFEE 358
EKWLPRRK MYSLA RFPKLLSFFYRKSFLPE+H+ IDK S+S GKKD+++ +EP FEE
Sbjct: 181 EKWLPRRKMMYSLARRFPKLLSFFYRKSFLPEQHDEIDKLLSVSPGKKDKLMTEEPEFEE 360
Query: 359 YWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIEELHVHKKCQTGGLLLWLKSMYGQAEC 418
+WQRD+EESVRQGN++PFIEEA+LQVS W F+I+ELHV KKCQT G+LLWLKSMY QA+C
Sbjct: 361 FWQRDVEESVRQGNIRPFIEEAVLQVSNWGFDIKELHVQKKCQTRGILLWLKSMYSQADC 540
Query: 419 ELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPEAVIHKLPNEGHFSYFFFCDECHKQIF 478
ELAG+LG HIWQGLDDR+VPPS+ EYIERVLPEA IHKLPNEGHFSYF+FCD+CH+QIF
Sbjct: 541 ELAGFLGLKHIWQGLDDRVVPPSVMEYIERVLPEAAIHKLPNEGHFSYFYFCDQCHRQIF 720
Query: 479 STLFGTPQGPFERQEETMLEKNTEDALVSV 508
+TLFGTPQGP ERQEET E + V V
Sbjct: 721 ATLFGTPQGPVERQEETATVLEEEVSQVPV 810
>TC231397 weakly similar to GB|AAP21157.1|30102478|BT006349
At5g02970/F9G14_280 {Arabidopsis thaliana;} , partial
(33%)
Length = 830
Score = 271 bits (692), Expect = 7e-73
Identities = 123/218 (56%), Positives = 167/218 (76%), Gaps = 4/218 (1%)
Frame = +3
Query: 289 MTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKHERIDKQFSLSLGKKDE 348
+TK+E +RTW KW +RK+MY LA RFP+LL+FFYR+SFL KH +ID+ SLSLG +D+
Sbjct: 21 ITKEERRRTWNKWTRKRKFMYFLARRFPRLLAFFYRRSFLSGKHGQIDRWLSLSLGNRDK 200
Query: 349 ILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIEELHVHKKCQTGGLLLW 408
L+++P + E+WQRD+EES RQ N+KPF+EE LQV+ W F++ +L + K+ Q+ LL W
Sbjct: 201 ALMEDPIYGEFWQRDVEESNRQRNVKPFMEEDALQVANWGFSLSDLKLQKRKQSSNLLSW 380
Query: 409 LKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPEAVIHKLPNEGHFSYFF 468
LKSM+ + E E G+LG IHIWQG+DD++VPPSMT+++ R+LP A +HKLP EGHF+Y +
Sbjct: 381 LKSMFTETE-EYMGFLGPIHIWQGMDDKVVPPSMTDFVHRLLPGAAVHKLPYEGHFTYIY 557
Query: 469 FCDECHKQIFSTLFGTPQGP----FERQEETMLEKNTE 502
FC ECH+QIF+TLFGTPQGP E + T+ E N E
Sbjct: 558 FCHECHRQIFTTLFGTPQGPLSISIEVDQATLEETNIE 671
>TC211233 weakly similar to GB|AAP21157.1|30102478|BT006349
At5g02970/F9G14_280 {Arabidopsis thaliana;} , partial
(7%)
Length = 497
Score = 152 bits (383), Expect = 5e-37
Identities = 81/124 (65%), Positives = 91/124 (73%), Gaps = 3/124 (2%)
Frame = +2
Query: 1 MAEARGTTWWSEELASLMENS---LPESSTTTSFEDVRKSSSSEEVEVGESLKEHAVGFL 57
MAE RG W+EELASL+E+S S TSFE S S GESLKE A+GF+
Sbjct: 140 MAEVRGRDTWTEELASLIEDSGVRYGGDSPPTSFEVHAPQSES-----GESLKEQALGFV 304
Query: 58 MAWCEILMELGRGCRDILQQNFFNEDSYLVQKLRGPCSKLSKRLSFLNDFLPEDRDPLLA 117
MAWCEILMELGRG RDIL+QN NEDSY+V+K GPCSK+SKRL FLNDFLPEDRDP A
Sbjct: 305 MAWCEILMELGRGFRDILRQNLINEDSYVVRKFGGPCSKVSKRLRFLNDFLPEDRDPFHA 484
Query: 118 WSIV 121
WS+V
Sbjct: 485 WSVV 496
>TC222551 similar to GB|AAS49080.1|44917511|BT011717 At3g44520 {Arabidopsis
thaliana;} , partial (92%)
Length = 639
Score = 117 bits (293), Expect = 1e-26
Identities = 64/187 (34%), Positives = 109/187 (58%), Gaps = 4/187 (2%)
Frame = +3
Query: 147 VRMHPPIASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDY 206
V + PP R+ L DGRY+AY+++GVP +A+ S++ H F SS+ + L+++
Sbjct: 81 VPVSPP---RVRLRDGRYLAYREKGVPKDQAKHSIIIVHGFGSSKDMNFLAPQ-ELIDEL 248
Query: 207 GVRLVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLK 266
G+ ++ YD G+GESDP+P R+ S A+D+ L D + + KF+++ S G W+
Sbjct: 249 GIYILQYDRAGYGESDPNPKRSLKSEALDIEELADLLQIGSKFYLIGVSMGSYATWSCFN 428
Query: 267 YIPERIAGAAMLAPMVS----PYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFF 322
YIP R+AG AM+AP+++ + + K++ +R KW M+ Y FP+LL ++
Sbjct: 429 YIPNRLAGVAMIAPVINYLWPSFPESLIKEDYRRKLIKW-----SMWFANY-FPRLLYWW 590
Query: 323 YRKSFLP 329
+ +LP
Sbjct: 591 VTQKWLP 611
>TC222969
Length = 323
Score = 114 bits (285), Expect = 1e-25
Identities = 62/112 (55%), Positives = 75/112 (66%), Gaps = 10/112 (8%)
Frame = +2
Query: 2 AEARGTTWWSEELASLMENS----------LPESSTTTSFEDVRKSSSSEEVEVGESLKE 51
A+ RG W+EEL SL+E+S ++TTTSFE S S GES KE
Sbjct: 2 ADVRGRDTWTEELPSLIEDSGVLYGGDPTTTTTTTTTTSFEVYAPHSES-----GESFKE 166
Query: 52 HAVGFLMAWCEILMELGRGCRDILQQNFFNEDSYLVQKLRGPCSKLSKRLSF 103
HA+GF+MAWCEILMELGRG RDIL+QN NEDS++V+K GPCS +SKRL F
Sbjct: 167 HALGFVMAWCEILMELGRGFRDILRQNLMNEDSFVVRKFGGPCSTVSKRLRF 322
>TC232456
Length = 746
Score = 112 bits (280), Expect = 4e-25
Identities = 55/134 (41%), Positives = 85/134 (63%), Gaps = 1/134 (0%)
Frame = +1
Query: 151 PPIAS-RILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVR 209
P +AS R+ L DGR++AY++ GVP AR+ ++ H + SS+ +P V L+ED G+
Sbjct: 271 PEVASPRVKLSDGRHLAYREFGVPKEEARYKIIVIHGYDSSKDTSLP-VSQELVEDLGIY 447
Query: 210 LVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIP 269
+ +D G+GESDPH R+ S A D+ L D + + KF+++ S G W+ LKYIP
Sbjct: 448 FLHFDRAGYGESDPHSLRSVKSEAYDIQELADKLEIGHKFYIIGMSMGGYPVWSCLKYIP 627
Query: 270 ERIAGAAMLAPMVS 283
R++GAA++AP +S
Sbjct: 628 HRLSGAALVAPFIS 669
>TC214621 similar to UP|Q84VY8 (Q84VY8) At2g36290, partial (62%)
Length = 1468
Score = 83.6 bits (205), Expect(2) = 8e-23
Identities = 60/246 (24%), Positives = 110/246 (44%)
Frame = +2
Query: 218 FGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIPERIAGAAM 277
+GESDP P+R S A+D+ L D + + KF+V+ S G W LKYIP R+AGA +
Sbjct: 494 YGESDPDPNRTLKSLALDIEQLADHLGLGSKFYVVGVSMGGQVVWNCLKYIPNRLAGAVL 673
Query: 278 LAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKHERIDK 337
++P+V+ + + + + K ++ +A+ P L ++ + + P
Sbjct: 674 ISPVVNYWWPGLPANLTTEAFSKKKLEDRWALRVAHYIPWLTYWWNTQRWFPASTAIAHS 853
Query: 338 QFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIEELHVH 397
+LS +D+ LV + + + + + + QG+ + + + W+++ +L
Sbjct: 854 PDNLS--HQDKELVPKMSNRKSYVAQVRQ---QGDYETLHRDLNIGFGNWEYSPLDL--- 1009
Query: 398 KKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPEAVIHK 457
E G +H+W G +D MVP ++ YI + LP H+
Sbjct: 1010-------------------ENPFPNNEGSVHLWHGDEDLMVPVTLQRYIAQKLPWIHYHE 1132
Query: 458 LPNEGH 463
L GH
Sbjct: 1133LQGSGH 1150
Score = 42.0 bits (97), Expect(2) = 8e-23
Identities = 24/73 (32%), Positives = 42/73 (56%), Gaps = 3/73 (4%)
Frame = +3
Query: 151 PPI-ASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPG--VKASLLEDYG 207
PPI A RI L DGR++AY++ GVP A++ +++ H+F R + + ++ +G
Sbjct: 288 PPITAPRIKLRDGRHLAYKEHGVPKDAAKYKIISVHAFDCCRHDTVVANTLSPDVVRSWG 467
Query: 208 VRLVTYDLPGFGE 220
L+ + PG G+
Sbjct: 468 STLI-FHRPGMGK 503
>TC214551 similar to UP|Q9SJM9 (Q9SJM9) Expressed protein, partial (34%)
Length = 662
Score = 81.3 bits (199), Expect = 1e-15
Identities = 40/105 (38%), Positives = 66/105 (62%), Gaps = 3/105 (2%)
Frame = +2
Query: 151 PPI-ASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPG--VKASLLEDYG 207
PPI A RI L DGR++AY++ GVP A++ +++ H F S R + + ++E+ G
Sbjct: 338 PPITAPRIKLRDGRHLAYKEHGVPKDAAKYKIISVHGFNSCRHDTVVANTLSPDVVEELG 517
Query: 208 VRLVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVL 252
+ +V++D PG+GESDP P+R S +D+ L D + + KF+V+
Sbjct: 518 LYIVSFDRPGYGESDPDPNRTLKSITLDIQELADQLGLGSKFYVV 652
>BM308920
Length = 420
Score = 56.2 bits (134), Expect = 3e-08
Identities = 25/59 (42%), Positives = 35/59 (58%)
Frame = +3
Query: 213 YDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIPER 271
+D G+GESDPH R+ S A D+ L D + + KF+++ S G W+ LKYIP R
Sbjct: 3 FDRAGYGESDPHSLRSVKSEAYDIQELADKLEIGHKFYIIGMSMGGYPVWSCLKYIPHR 179
>TC219938
Length = 819
Score = 53.1 bits (126), Expect = 3e-07
Identities = 46/197 (23%), Positives = 85/197 (42%), Gaps = 4/197 (2%)
Frame = +1
Query: 271 RIAGAAMLAPMVS----PYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKS 326
R+AG AM+AP+++ + + K++ +R KW M+ Y FP+LL ++ +
Sbjct: 97 RLAGVAMIAPVINYLWPSFPESLIKEDYRRKLIKWS-----MWFANY-FPRLLYWWVTQK 258
Query: 327 FLPEKHERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSR 386
+LP + I+K + + +IL P F + L E V L+ + ++
Sbjct: 259 WLPS-NSVIEKNPAFFNKRDIDILETIPGFPMLTKNKLREQVVFDTLRG---DWMVAFGN 426
Query: 387 WDFNIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYI 446
W+F+ +L HIWQG +D++VP + ++
Sbjct: 427 WEFDPLKL----------------------SNPFPDNRSSAHIWQGYEDKVVPSQIQRFV 540
Query: 447 ERVLPEAVIHKLPNEGH 463
+ LP H++P+ GH
Sbjct: 541 TQKLPWIQYHEVPDGGH 591
>TC233192 weakly similar to UP|Q84VY8 (Q84VY8) At2g36290, partial (12%)
Length = 232
Score = 46.2 bits (108), Expect = 3e-05
Identities = 21/53 (39%), Positives = 31/53 (57%)
Frame = +3
Query: 425 GRIHIWQGLDDRMVPPSMTEYIERVLPEAVIHKLPNEGHFSYFFFCDECHKQI 477
G +HIWQG +DR++P ++ YI LP H+L + GH + F +EC I
Sbjct: 30 GSVHIWQGFEDRIIPYTLNRYISDRLPWIRYHELAHAGHL-FLFKKNECESII 185
>TC214509 similar to UP|Q84VY8 (Q84VY8) At2g36290, partial (31%)
Length = 815
Score = 42.4 bits (98), Expect = 5e-04
Identities = 28/84 (33%), Positives = 38/84 (44%)
Frame = +1
Query: 425 GRIHIWQGLDDRMVPPSMTEYIERVLPEAVIHKLPNEGHFSYFFFCDECHKQIFSTLFGT 484
G +H+WQG +D MVP ++ YI + LP H+L GH F D I +L
Sbjct: 298 GSVHLWQGDEDMMVPVTLQRYIAQNLPWINYHELQGSGHI--FAHADGMSDTIIKSLLRA 471
Query: 485 PQGPFERQEETMLEKNTEDALVSV 508
F T+ EK LV+V
Sbjct: 472 K*ASF--MLVTVCEKLAASILVTV 537
>BU089844 similar to PIR|A86217|A862 protein T23G18.18 [imported] -
Arabidopsis thaliana, partial (12%)
Length = 365
Score = 42.4 bits (98), Expect = 5e-04
Identities = 20/45 (44%), Positives = 31/45 (68%)
Frame = +3
Query: 147 VRMHPPIASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSR 191
V + PP R+ L DGRY+AY+++GVP +A+ S++ H F SS+
Sbjct: 165 VPVSPP---RVRLRDGRYLAYREKGVPKDQAKHSIIIVHGFGSSK 290
>BQ079462 similar to PIR|A86217|A862 protein T23G18.18 [imported] -
Arabidopsis thaliana, partial (12%)
Length = 423
Score = 42.4 bits (98), Expect = 5e-04
Identities = 20/45 (44%), Positives = 31/45 (68%)
Frame = +3
Query: 147 VRMHPPIASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSR 191
V + PP R+ L DGRY+AY+++GVP +A+ S++ H F SS+
Sbjct: 111 VPVSPP---RVRLRDGRYLAYREKGVPKDQAKHSIIIVHGFGSSK 236
>AW704298 similar to PIR|A86217|A862 protein T23G18.18 [imported] -
Arabidopsis thaliana, partial (12%)
Length = 419
Score = 41.2 bits (95), Expect = 0.001
Identities = 20/45 (44%), Positives = 31/45 (68%)
Frame = +2
Query: 147 VRMHPPIASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSR 191
V + PP RI L DGR++AY+++GVP +A+ S++ H F SS+
Sbjct: 167 VPVSPP---RIKLRDGRFLAYREKGVPKDQAKHSIIIVHGFGSSK 292
>BE059550
Length = 357
Score = 39.7 bits (91), Expect = 0.003
Identities = 17/68 (25%), Positives = 38/68 (55%)
Frame = +3
Query: 262 WASLKYIPERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSF 321
W LK+IP R+AGA ++ P+V+ + ++ + + + K ++ +A+ FP L +
Sbjct: 9 WGCLKFIPHRLAGATLMTPVVNYWWHNLPLNMTTKAYYKQPKHDQWALRVAHYFPWLTYW 188
Query: 322 FYRKSFLP 329
++ + + P
Sbjct: 189 WFTQEWFP 212
>TC226960 similar to UP|Q9ZP87 (Q9ZP87) Epoxide hydrolase, partial (72%)
Length = 1201
Score = 39.3 bits (90), Expect = 0.004
Identities = 28/89 (31%), Positives = 45/89 (50%), Gaps = 4/89 (4%)
Frame = +1
Query: 207 GVRLVTYDLPGFGESDPHP---SRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWA 263
G R V++D G+G SDP P +++ D+LH++DA+ ++ K +++ G A+
Sbjct: 208 GFRAVSFDYRGYGLSDPPPEPDKTSWSDLLSDLLHILDALALS-KVFLVGKDFGARPAYL 384
Query: 264 SLKYIPERIAGAAML-APMVSPYESHMTK 291
PER+ G L P V P S K
Sbjct: 385 FSILHPERVLGVVTLGVPYVPPGPSQYHK 471
>BI973866 weakly similar to GP|1354849|gb| epoxide hydrolase {Nicotiana
tabacum}, partial (33%)
Length = 420
Score = 38.5 bits (88), Expect = 0.007
Identities = 29/86 (33%), Positives = 43/86 (49%), Gaps = 4/86 (4%)
Frame = +1
Query: 203 LEDYGVRLVTYDLPGFGESDPHPSRN---FNSSAMDMLHLVDAVNVTDKFWVLCHSSGCI 259
L D G R V++D G+G SDP P N + D+LH++DA+ ++ K +++ G
Sbjct: 154 LADAGFRAVSFDYRGYGLSDPPPPGNKATWFDLLNDLLHILDALALS-KVFLVGKDFGAR 330
Query: 260 HAWASLKYIPERIAGAAML-APMVSP 284
A PER+ G L P V P
Sbjct: 331 PAHFFSILHPERVLGVVTLGVPYVPP 408
>TC233337 similar to GB|AAO63446.1|28951045|BT005382 At1g64670 {Arabidopsis
thaliana;} , partial (42%)
Length = 658
Score = 36.6 bits (83), Expect = 0.028
Identities = 37/123 (30%), Positives = 52/123 (42%), Gaps = 2/123 (1%)
Frame = +2
Query: 185 HSFLSSRLAGIPGVKASLLEDYGVRLVTYDLPGFGESD-PHPSRNFNSSAMDMLHL-VDA 242
H F+SS L V +L + RL DL GFG S P+ S ++M+ V
Sbjct: 116 HGFISSSLFWSETVFPNLSKSC-YRLFAVDLLGFGRSPKPNSSLYTLREHLEMIERSVLE 292
Query: 243 VNVTDKFWVLCHSSGCIHAWASLKYIPERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWL 302
+ F ++ HS GCI A A P+ + +LAP P T+ + K
Sbjct: 293 AHKVKSFHIVAHSLGCILALALAVKHPQSVKSLTLLAPPFYPVPKGETQ-ATQYVMRKVA 469
Query: 303 PRR 305
PRR
Sbjct: 470 PRR 478
>BE210363
Length = 400
Score = 34.3 bits (77), Expect = 0.14
Identities = 18/44 (40%), Positives = 26/44 (58%), Gaps = 3/44 (6%)
Frame = +3
Query: 203 LEDYGVRLVTYDLPGFGESDPHPSRN---FNSSAMDMLHLVDAV 243
L D G R V++D G+G SDP P N + D+LH++DA+
Sbjct: 255 LADAGFRAVSFDYRGYGLSDPPPPGNKATWFDLLNDLLHILDAL 386
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.136 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,761,284
Number of Sequences: 63676
Number of extensions: 400206
Number of successful extensions: 1904
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 1889
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1898
length of query: 512
length of database: 12,639,632
effective HSP length: 101
effective length of query: 411
effective length of database: 6,208,356
effective search space: 2551634316
effective search space used: 2551634316
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC124963.7


