

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124963.21 + phase: 0 /pseudo
(739 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
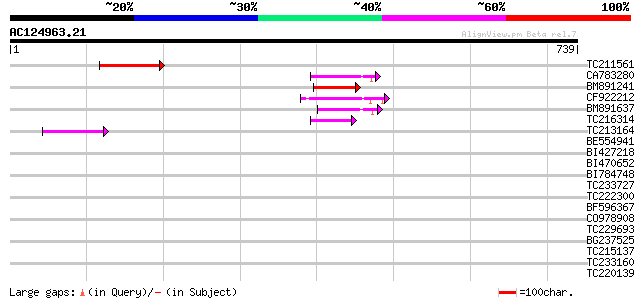
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 85 1e-16
CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polym... 79 9e-15
BM891241 76 6e-14
CF922212 69 1e-11
BM891637 66 5e-11
TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {A... 57 4e-08
TC213164 52 7e-07
BE554941 39 0.011
BI427218 39 0.011
BI470652 similar to PIR|T17102|T171 RNA-directed DNA polymerase ... 38 0.014
BI784748 36 0.054
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 36 0.071
TC222300 similar to GB|AAP37786.1|30725528|BT008427 At1g51560 {A... 35 0.092
BF596367 33 0.46
CO978908 33 0.60
TC229693 weakly similar to GB|CAA32948.1|1868|SSACROSI preproacr... 32 1.3
BG237525 homologue to SP|P13362|ACT1_ Actin 1. [Rice] {Oryza sat... 31 2.3
TC215137 30 5.1
TC233160 similar to UP|Q8S976 (Q8S976) Auxin response factor 10 ... 29 6.6
TC220139 similar to UP|FKB7_WHEAT (Q43207) 70 kDa peptidylprolyl... 29 6.6
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 85.1 bits (209), Expect = 1e-16
Identities = 42/85 (49%), Positives = 60/85 (70%)
Frame = +1
Query: 118 LDGILVANEVVDDAHRCKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKEC 177
L L+ANEV+D+A R K L+FKVD+E+AYD V W++L +M++M F K I+EC
Sbjct: 1 LHSALIANEVIDEAKRSNKSCLVFKVDYEKAYDSVSWNFLMYMMWRMDFSPKWIKWIEEC 180
Query: 178 IGTATTSMLVNGSPTDEFPLKRGCK 202
+ +A+ S+LVNGSPT EF +RG +
Sbjct: 181 VKSASISVLVNGSPTAEFLPQRGLR 255
Score = 33.1 bits (74), Expect = 0.46
Identities = 12/28 (42%), Positives = 17/28 (59%)
Frame = +1
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGG 420
F G + KK++W+ W +CL KE GG
Sbjct: 823 FLWGGGPDQKKISWIRWEKLCLPKERGG 906
>CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polymerase
homolog T24M8.8 - Arabidopsis thaliana, partial (4%)
Length = 456
Score = 78.6 bits (192), Expect = 9e-15
Identities = 37/94 (39%), Positives = 54/94 (57%), Gaps = 3/94 (3%)
Frame = +2
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRV 452
F G + +K+ WV+W VCL K GG G++ +R FN+ LLGKW W L ++ W +V
Sbjct: 62 FLWGGEADSRKIAWVNWKTVCLPKAKGGLGIKDLRTFNTTLLGKWRWDLFYIQQEPWAKV 241
Query: 453 LSARYNEEGGRLLDGGRS---TSAWWRVIATLQR 483
L ++Y G R L+ G S SAWW+ + Q+
Sbjct: 242 LQSKYG--GWRALEEGSSGSKDSAWWKDLIKTQQ 337
>BM891241
Length = 407
Score = 75.9 bits (185), Expect = 6e-14
Identities = 29/61 (47%), Positives = 42/61 (68%)
Frame = -2
Query: 397 GSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRVLSAR 456
G +HKK+ WV W VCL K GG G++ I +FN ALLGKW W L +++ LW R+++++
Sbjct: 403 GDHDHKKIPWVKWEVVCLPKTKGGLGIKDISKFNIALLGKWIWALASDQQQLWARIINSK 224
Query: 457 Y 457
Y
Sbjct: 223 Y 221
>CF922212
Length = 445
Score = 68.6 bits (166), Expect = 1e-11
Identities = 46/123 (37%), Positives = 61/123 (49%), Gaps = 7/123 (5%)
Frame = -2
Query: 379 QVLSPLLNLF*NVFFFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWC 438
+VL L+ L +F G KK+ WV+W+ VCL KE G G R +R+FN ALLGK
Sbjct: 432 KVLDKLVRL--QQWFLWGGGLGQKKIAWVNWDSVCLPKEKEGLGNRDLRKFNYALLGKRR 259
Query: 439 WQLLIEKESLWYRVLSARYNEEGGRLLDGGR---STSAWWRVIATLQRE----EWFNSCR 491
W L + L RVL ++Y R LD R S S WW+ I+ + WF
Sbjct: 258 WNLFHHQGELGARVLDSKYKR--WRNLDEERRVKSESFWWQEISFITHSTEDGSWFEKRT 85
Query: 492 EIG 494
+ G
Sbjct: 84 KTG 76
>BM891637
Length = 427
Score = 66.2 bits (160), Expect = 5e-11
Identities = 31/87 (35%), Positives = 48/87 (54%), Gaps = 3/87 (3%)
Frame = -1
Query: 402 KKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRVLSARYNEEG 461
KK+ W+ W C S +VGG G++ I+ N+ALL KW W + + LW R+L ++Y +G
Sbjct: 418 KKIAWISWRQCCASGDVGGLGIKDIKILNNALLIKWKWLMFHQPHQLWNRILISKY--KG 245
Query: 462 GRLLDGGRST---SAWWRVIATLQREE 485
R LD G S WW + + + +
Sbjct: 244 WRGLDQGPQKYYFSPWWADLRAINQHQ 164
>TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {Arabidopsis
thaliana;} , partial (18%)
Length = 759
Score = 56.6 bits (135), Expect = 4e-08
Identities = 22/60 (36%), Positives = 33/60 (54%)
Frame = +2
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRV 452
F G+ +H ++ WV W +C K GG G++ + FN+AL G+W W L LW R+
Sbjct: 119 FLWGGTQHHNRIPWVKWADICNPKIDGGLGIKDLSNFNAALRGRWIWGLASNHNQLWARL 298
>TC213164
Length = 446
Score = 52.4 bits (124), Expect = 7e-07
Identities = 30/88 (34%), Positives = 52/88 (59%), Gaps = 1/88 (1%)
Frame = +3
Query: 43 LVNFIGMGGLPKESIALSLLLFLRLRAR-RLNDFCLISLVESMYKILAKVLANRLRYVIG 101
L F G K S A L L ++ LN++ ISL+ +YKI+AK+L+ R + V+
Sbjct: 159 LDEFYANGVFLKRSNAFFLALIPKVHDP*SLNEYKPISLIGCIYKIVAKLLSGRHKKVLP 338
Query: 102 MVISDSQSTFVKGRQILDGILVANEVVD 129
+I + Q+ F++GR L +++ANE+++
Sbjct: 339 TIIDERQTVFMEGRHTLHNVVIANEIME 422
>BE554941
Length = 383
Score = 38.5 bits (88), Expect = 0.011
Identities = 26/92 (28%), Positives = 39/92 (42%), Gaps = 6/92 (6%)
Frame = +1
Query: 422 GVRRIREFNSALLGKWCWQLLIEKESLWYRVLSARYNEEGGRLLDGGRSTSAWWRVIATL 481
G++ + FN +LL K L+E +++W +L Y TS WWR I +
Sbjct: 7 GIKNLSIFNISLLVKRR*HFLVETKAIWAPILKFHYGSLDLSNTGSASRTSQWWRDILRI 186
Query: 482 ----QREEWFNS--CREIGGEWERNILLVRCL 507
+ W S CR+IG + RCL
Sbjct: 187 GNCQYQPGWLTSTFCRKIGDRKDTLFWRHRCL 282
>BI427218
Length = 421
Score = 38.5 bits (88), Expect = 0.011
Identities = 22/69 (31%), Positives = 30/69 (42%), Gaps = 5/69 (7%)
Frame = +1
Query: 435 GKWCWQLLIEKESLWYRVLSARYN-----EEGGRLLDGGRSTSAWWRVIATLQREEWFNS 489
GKW W K LW R+L ++Y EE R G SAWW+ I + S
Sbjct: 1 GKWKWSFFYNKGELWARILDSKYGDWRHLEEPTR----GNRASAWWKDINIIDPSGADGS 168
Query: 490 CREIGGEWE 498
+ G +W+
Sbjct: 169 LFDKGIKWK 195
>BI470652 similar to PIR|T17102|T171 RNA-directed DNA polymerase (EC
2.7.7.49) (clone AmLi2) - garden snapdragon (fragment),
partial (22%)
Length = 111
Score = 38.1 bits (87), Expect = 0.014
Identities = 16/28 (57%), Positives = 22/28 (78%)
Frame = +1
Query: 175 KECIGTATTSMLVNGSPTDEFPLKRGCK 202
KEC+ +A+ S+LVNGSPT+EF RG +
Sbjct: 1 KECLSSASISILVNGSPTEEFKPXRGLR 84
>BI784748
Length = 259
Score = 36.2 bits (82), Expect = 0.054
Identities = 14/33 (42%), Positives = 25/33 (75%)
Frame = +2
Query: 108 QSTFVKGRQILDGILVANEVVDDAHRCKKELLL 140
QS F++GR +L +++ANEV+D+A R +K ++
Sbjct: 14 QSAFIQGRHMLHSMMIANEVIDEAKRGQKPSIM 112
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 35.8 bits (81), Expect = 0.071
Identities = 20/64 (31%), Positives = 33/64 (51%)
Frame = -3
Query: 139 LLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLK 198
L K+D +A+D +D +L V+ K G+ I+ + +A S+ VNG P F +
Sbjct: 765 LALKIDIVKAFDTLDRDFLISVLNKFGYSHTFCNWIRVILLSAKLSISVNGEPVGFFSCQ 586
Query: 199 RGCK 202
RG +
Sbjct: 585 RGVR 574
>TC222300 similar to GB|AAP37786.1|30725528|BT008427 At1g51560 {Arabidopsis
thaliana;} , partial (37%)
Length = 801
Score = 35.4 bits (80), Expect = 0.092
Identities = 12/35 (34%), Positives = 22/35 (62%)
Frame = +1
Query: 470 STSAWWRVIATLQREEWFNSCREIGGEWERNILLV 504
S ++WW+ + TL + + N CR++G W R I+ +
Sbjct: 637 SQASWWKQLLTLTKRSFVNMCRDVGYYWLRIIVYI 741
>BF596367
Length = 419
Score = 33.1 bits (74), Expect = 0.46
Identities = 15/34 (44%), Positives = 24/34 (70%)
Frame = +1
Query: 424 RRIREFNSALLGKWCWQLLIEKESLWYRVLSARY 457
RRI+ F+ A+L K W+L+ K++L VL+A+Y
Sbjct: 58 RRIKAFHKAMLAKQMWRLIDNKDALVT*VLTAKY 159
>CO978908
Length = 782
Score = 32.7 bits (73), Expect = 0.60
Identities = 14/43 (32%), Positives = 22/43 (50%), Gaps = 3/43 (6%)
Frame = +1
Query: 437 WCWQLLIEKESLWYRVLSARYNEEGGRLLDGG---RSTSAWWR 476
W W L +++ LW R+++++Y G G R S WWR
Sbjct: 604 WIWALASDQQQLWARIINSKYG--GWEEFQSGRDKRGCSHWWR 726
>TC229693 weakly similar to GB|CAA32948.1|1868|SSACROSI preproacrosin {Sus
scrofa;} , partial (8%)
Length = 520
Score = 31.6 bits (70), Expect = 1.3
Identities = 20/50 (40%), Positives = 30/50 (60%), Gaps = 1/50 (2%)
Frame = +2
Query: 340 KLDCQGGKLNIYLLVVIWFC*NLSCHLSL-FMLFLSSRLLQVLSPLLNLF 388
+L G L++ L++ FC* CHLSL F L LS+R +Q+ S + +F
Sbjct: 314 RLHASTGSLSLLLVLYSTFC*CFQCHLSLPFFLSLSNR-VQLKSVCVRVF 460
>BG237525 homologue to SP|P13362|ACT1_ Actin 1. [Rice] {Oryza sativa},
partial (20%)
Length = 436
Score = 30.8 bits (68), Expect = 2.3
Identities = 19/65 (29%), Positives = 34/65 (52%), Gaps = 5/65 (7%)
Frame = -1
Query: 327 VVCLFGNH*LGGSKLDCQGGKLNIYLLVVIWFC*NLSCHLS-----LFMLFLSSRLLQVL 381
+ CL H L G++ CQ G N ++ +W+C +L+ H + + +L LS+ ++
Sbjct: 196 IFCLCNPHLLEGAE-GCQNGASNPDTVLPLWWCHHLNFHAAWC*CVISLLILSAMPGNIV 20
Query: 382 SPLLN 386
P LN
Sbjct: 19 EPPLN 5
>TC215137
Length = 1003
Score = 29.6 bits (65), Expect = 5.1
Identities = 11/18 (61%), Positives = 13/18 (72%)
Frame = -2
Query: 694 RCLRSLYSVHFWWRRISE 711
R LR L+ + FWWRR SE
Sbjct: 333 RALRWLFMIDFWWRRCSE 280
>TC233160 similar to UP|Q8S976 (Q8S976) Auxin response factor 10 (Fragment),
partial (26%)
Length = 658
Score = 29.3 bits (64), Expect = 6.6
Identities = 24/82 (29%), Positives = 34/82 (41%), Gaps = 7/82 (8%)
Frame = +1
Query: 401 HKKVTWVDW--NFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWY-----RVL 453
H K T+VD+ + C S+E +G+ + F G CW E+ +LW+ RV
Sbjct: 100 HAKATFVDYWVEYFCESQEAFCWGLNCV--FEGRKWGFVCWDSTSEEGNLWWT*DFLRVE 273
Query: 454 SARYNEEGGRLLDGGRSTSAWW 475
S R S S WW
Sbjct: 274 SCRRK----------LSYSLWW 309
>TC220139 similar to UP|FKB7_WHEAT (Q43207) 70 kDa peptidylprolyl isomerase
(Peptidyl-prolyl cis-trans isomerase) (PPIase)
(Rotamase) , partial (29%)
Length = 722
Score = 29.3 bits (64), Expect = 6.6
Identities = 11/24 (45%), Positives = 15/24 (61%)
Frame = -2
Query: 193 DEFPLKRGCKAIRIPLSFFSWLQR 216
D FP +RGC+++ L FF W R
Sbjct: 145 DPFPRRRGCRSLLPSLPFFHWKTR 74
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.350 0.157 0.564
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,393,200
Number of Sequences: 63676
Number of extensions: 712395
Number of successful extensions: 7088
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 5201
Number of HSP's successfully gapped in prelim test: 206
Number of HSP's that attempted gapping in prelim test: 1673
Number of HSP's gapped (non-prelim): 5682
length of query: 739
length of database: 12,639,632
effective HSP length: 104
effective length of query: 635
effective length of database: 6,017,328
effective search space: 3821003280
effective search space used: 3821003280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC124963.21


