

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
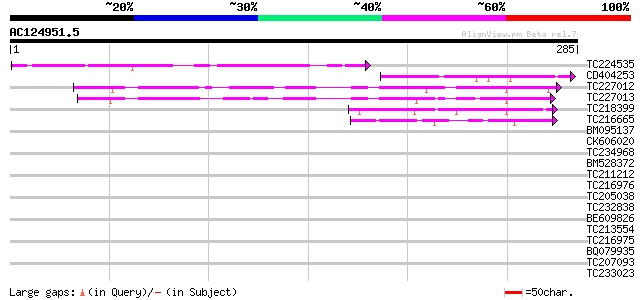
Query= AC124951.5 + phase: 0
(285 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC224535 GB|CAB64866.1|6687237|CEL243905 SF1 protein {Caenorhabd... 105 3e-23
CD404253 74 8e-14
TC227012 70 1e-12
TC227013 similar to UP|GAC1_YEAST (P28006) Protein phosphatase 1... 64 6e-11
TC218399 UP|PA2X_HUMAN (O15496) Group X secretory phospholipase ... 50 1e-06
TC216665 similar to UP|Q7XHJ1 (Q7XHJ1) Pheromone receptor-like p... 41 7e-04
BM095137 34 0.066
CK606020 33 0.15
TC234968 similar to UP|Q9IPQ8 (Q9IPQ8) EBNA-1, partial (5%) 33 0.19
BM528372 32 0.33
TC211212 similar to UP|Q40238 (Q40238) Homeobox transcription fa... 31 0.56
TC216976 similar to UP|Q6H974 (Q6H974) SEU1 protein, partial (6%) 31 0.73
TC205038 similar to UP|Q94II3 (Q94II3) ERD3 protein, partial (48%) 30 0.96
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 30 0.96
BE609826 30 1.6
TC213554 weakly similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protei... 30 1.6
TC216975 similar to UP|Q6H974 (Q6H974) SEU1 protein, partial (28%) 30 1.6
BQ079935 30 1.6
TC207093 29 2.1
TC233023 similar to UP|O48652 (O48652) 6-4 photolyase, partial (... 29 2.1
>TC224535 GB|CAB64866.1|6687237|CEL243905 SF1 protein {Caenorhabditis
elegans;} , partial (3%)
Length = 502
Score = 105 bits (261), Expect = 3e-23
Identities = 73/187 (39%), Positives = 100/187 (53%), Gaps = 7/187 (3%)
Frame = +2
Query: 2 QNQQQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQND-DFEFVAFRSHR 60
+ Q+ EE S M+S +CPSFS YSSN ++D+A QV ND +ND DFEFVAFR
Sbjct: 11 KKMQKVEE--SVMESLVCPSFSAYSSNTLDDIADQVI-RNDVRFQENDTDFEFVAFRKVA 181
Query: 61 ------RNAADVSPIFNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILY 114
NA PIF+RD ++ G+ K + +G D A I
Sbjct: 182 DGVFVDNNATPAFPIFDRDLATAAEEDS----------GEGKRRASG---EEDDVAAIQI 322
Query: 115 SLKKLFIGNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSN 174
+L KL + + +SSEVED+L+++P +YC WTP+ K SP +T C+KS
Sbjct: 323 TLGKLLMDDSSASCSSSEVEDELENVPPGTYCVWTPR--------KASPAPAT-PCRKSK 475
Query: 175 STGSSSN 181
STGSSS+
Sbjct: 476 STGSSSS 496
>CD404253
Length = 617
Score = 73.9 bits (180), Expect = 8e-14
Identities = 46/108 (42%), Positives = 65/108 (59%), Gaps = 10/108 (9%)
Frame = -3
Query: 187 WKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVVGKKIPAT----EKKTPA--T 240
WK L LLRRS S+GKES+ +TP+ + K G + GKK PA+ EK+ A
Sbjct: 615 WKLLDLLRRSNSEGKESVVFLTPSSVNSAK--KKGTKSETGKKSPASSGGGEKRIVAVPA 442
Query: 241 VSAMEVFYGR----KKETRVKSYLPYKKELIGFSVGFNANVGRGFPLH 284
VSA E Y R ++E + +SYLPY+++L+G V N ++G+ FPLH
Sbjct: 441 VSAHEALYVRNREMRREVKRRSYLPYRQDLVGLCVNLN-SMGKAFPLH 301
>TC227012
Length = 931
Score = 70.1 bits (170), Expect = 1e-12
Identities = 85/264 (32%), Positives = 114/264 (42%), Gaps = 19/264 (7%)
Frame = +3
Query: 33 VAQQVTNENDNSHSQNDD-------FEFVAFRSHRRNAADVSPIFNRDERRNSDAMDI-S 84
VA+ NEN+N N+D F FV S D SPI D N I
Sbjct: 177 VAKLKINENENEEKHNEDEVEEEEEFSFVLTNS------DGSPISAEDAFDNGHIRPIFP 338
Query: 85 ISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIGNEKEKWTSSEVEDDLDSIPAE- 143
I ++LL D+ NGG + L K+F+ + + SE + PAE
Sbjct: 339 IFNQDLLFSDD---YNGG--------DGLRLPIKVFVEHADVSPSPSEA-----TTPAEG 470
Query: 144 SYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKES 203
+YC W PK++ KSNSTG S W+F + RS SDGK++
Sbjct: 471 TYCEWNPKAAV-----------------KSNSTGFSK-----LWRFRDVKLRSNSDGKDA 584
Query: 204 LNMVT--PAKKENLKLNSGGNGNVVGKKIPATE-KKTPATVSAMEVFY----GRKKETRV 256
+ PA K K +S VV KK+ + K T A SA E Y RK+ +
Sbjct: 585 FVFLNHAPAAKPAEKASS-----VVVKKVEVKKGKTTTAAASAHEKHYVMSRARKESDKR 749
Query: 257 KSYLPYKKELIGF---SVGFNANV 277
KSYLPYK++L GF + G + NV
Sbjct: 750 KSYLPYKQDLFGFFANASGLSRNV 821
>TC227013 similar to UP|GAC1_YEAST (P28006) Protein phosphatase 1 regulatory
subunit GAC1, partial (3%)
Length = 1149
Score = 64.3 bits (155), Expect = 6e-11
Identities = 80/263 (30%), Positives = 110/263 (41%), Gaps = 23/263 (8%)
Frame = +2
Query: 35 QQVTNENDNSHSQND--------------DFEFVAFRSHRRNAADVSPIFNRDERRNSDA 80
++V NEN+ H++++ +F FV S D SPI D N
Sbjct: 191 EKVENENEEKHNEDEIEEEEEEEEEEEEEEFSFVLTNS------DGSPISADDAFDNGQI 352
Query: 81 MDI-SISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIGNEKEKWTSSEVEDDLDS 139
I I ++LL D+ D E L K+F+ K+++ S ++
Sbjct: 353 RPIFPIFNQDLLFSDDY-----------DGGEGLRQPIKVFV-EHKDEFLS-------EA 475
Query: 140 IPAE-SYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKS 198
PAE +YC W PK++ KSNSTG S W+F + RS S
Sbjct: 476 APAEGAYCEWNPKAAV-----------------KSNSTGFSK-----LWRFRDVKLRSNS 589
Query: 199 DGKES---LNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKTPATVSAMEVFY----GRK 251
DGK++ LN PAK K S VV KK T T A SA E Y RK
Sbjct: 590 DGKDAFVFLNHAAPAKPAE-KARS-----VVVKKGKTTT--TTAAASAHEKHYIMSRARK 745
Query: 252 KETRVKSYLPYKKELIGFSVGFN 274
+ + KSYLPYK+ ++GF N
Sbjct: 746 ESDKRKSYLPYKQNMLGFFANAN 814
>TC218399 UP|PA2X_HUMAN (O15496) Group X secretory phospholipase A2 precursor
(Phosphatidylcholine 2-acylhydrolase GX) (GX sPLA2)
(sPLA2-X) , partial (7%)
Length = 1360
Score = 50.1 bits (118), Expect = 1e-06
Identities = 43/122 (35%), Positives = 59/122 (48%), Gaps = 17/122 (13%)
Frame = +2
Query: 171 KKSN---STGSSSNTSSSRWKFLSLLRRSKSDGKE-----SLNMVTPAKKENLKLNSGGN 222
KK N S S S SS RW FL RSKS+G+ S +PA K+ K + GN
Sbjct: 614 KKENVDPSVSSGSGRSSKRWVFLRDFLRSKSEGRSNNKFWSTISFSPAAKDK-KPQNQGN 790
Query: 223 G-----NVVGKKIPATEKKTPATVSAMEVFY----GRKKETRVKSYLPYKKELIGFSVGF 273
G V GK KK S E+ Y + +E R K++LPY++ L+G +GF
Sbjct: 791 GPQKPKKVAGKPTNGIGKKRVPAASPHELHYKANRAQAEELRRKTFLPYRQGLLG-CLGF 967
Query: 274 NA 275
++
Sbjct: 968 SS 973
>TC216665 similar to UP|Q7XHJ1 (Q7XHJ1) Pheromone receptor-like protein
(Fragment), partial (42%)
Length = 1006
Score = 40.8 bits (94), Expect = 7e-04
Identities = 38/117 (32%), Positives = 58/117 (49%), Gaps = 13/117 (11%)
Frame = +2
Query: 172 KSNSTGSSSNTSSSRWKFLS-LLRRSKSDGKESLNMVTPAKK--------ENLKLNSGGN 222
+SN S S TS RW+ LL RS S+G+ S P +K E+ K ++ G
Sbjct: 518 QSNKADSLSKTSR-RWRLTDFLLFRSASEGRGSSK--DPLRKFPTFYKKPEDPKASNPG- 685
Query: 223 GNVVGKKIPATEKKTPATVSAMEVFYGRKK----ETRVKSYLPYKKELIGFSVGFNA 275
P T +K P +SA E+ Y RKK + + +++LPYK+ ++G GF +
Sbjct: 686 --------PKTRRKEP--LSAHELHYARKKAETEDLKKRTFLPYKQGILGRLAGFGS 826
>BM095137
Length = 428
Score = 34.3 bits (77), Expect = 0.066
Identities = 26/94 (27%), Positives = 41/94 (42%), Gaps = 12/94 (12%)
Frame = -2
Query: 146 CFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSN------------TSSSRWKFLSLL 193
C W P+ S+ P TS + KC + +T S+ + +++ + FLS L
Sbjct: 403 CIWNPRHVSI---PGTSASLGSTKCPEQTTTASNPSRLTESPSRLVLDSTTHSFPFLSTL 233
Query: 194 RRSKSDGKESLNMVTPAKKENLKLNSGGNGNVVG 227
S S+ SL +PA + S GNV+G
Sbjct: 232 TTSVSNWVNSLIPNSPANPSRNRSTSS*PGNVLG 131
>CK606020
Length = 340
Score = 33.1 bits (74), Expect = 0.15
Identities = 23/67 (34%), Positives = 32/67 (47%)
Frame = -3
Query: 152 SSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPAK 211
SS +S TS S+ S+S SSS++SSS S S SDG S ++ T
Sbjct: 332 SSPSTSSSSTSSSTSSSSSSSSSSISSSSSSSSSSSSSSSSSSSSSSDGSISSSLATGES 153
Query: 212 KENLKLN 218
++ LN
Sbjct: 152 GSDIGLN 132
Score = 29.6 bits (65), Expect = 1.6
Identities = 21/69 (30%), Positives = 34/69 (48%)
Frame = -3
Query: 139 SIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKS 198
S P+ S + +SS +S +S +S+ S+S+ SSS++SSS S L +S
Sbjct: 332 SSPSTSSSSTSSSTSSSSSSSSSSISSSSSSSSSSSSSSSSSSSSSSDGSISSSLATGES 153
Query: 199 DGKESLNMV 207
LN +
Sbjct: 152 GSDIGLNWI 126
>TC234968 similar to UP|Q9IPQ8 (Q9IPQ8) EBNA-1, partial (5%)
Length = 412
Score = 32.7 bits (73), Expect = 0.19
Identities = 22/83 (26%), Positives = 40/83 (47%), Gaps = 10/83 (12%)
Frame = +1
Query: 131 SEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNS----------TIKCKKSNSTGSSS 180
+E ED+ D + ++ P +++I+S + + ++ S+S SS+
Sbjct: 100 AENEDEEDHVHDKNVT--KPNQTNIISSSEAEKVEDGLGLGLETTPSVSGSASSSRSSSA 273
Query: 181 NTSSSRWKFLSLLRRSKSDGKES 203
SS RW FL RSKS+G+ +
Sbjct: 274 GRSSKRWVFLKDFLRSKSEGRSN 342
>BM528372
Length = 411
Score = 32.0 bits (71), Expect = 0.33
Identities = 15/31 (48%), Positives = 20/31 (64%)
Frame = +3
Query: 173 SNSTGSSSNTSSSRWKFLSLLRRSKSDGKES 203
S+S SS+ SS RW FL RSKS+G+ +
Sbjct: 273 SSSRSSSAGRSSKRWVFLKDFLRSKSEGRSN 365
>TC211212 similar to UP|Q40238 (Q40238) Homeobox transcription factor Hox7
(Fragment), partial (15%)
Length = 483
Score = 31.2 bits (69), Expect = 0.56
Identities = 27/90 (30%), Positives = 43/90 (47%)
Frame = +3
Query: 177 GSSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVVGKKIPATEKK 236
G S S + KF + ++ ++G S +TP + E+ +L S N V +K + K
Sbjct: 3 GGPSLFSQATEKFCTFRPKTLTEGCCSPQFLTPME-ESSELQSEEN-KVSNEKNKKRKLK 176
Query: 237 TPATVSAMEVFYGRKKETRVKSYLPYKKEL 266
TPA + A+E FY K + L +EL
Sbjct: 177 TPAQLKALENFYNEHKYPTEEMKLVLAEEL 266
>TC216976 similar to UP|Q6H974 (Q6H974) SEU1 protein, partial (6%)
Length = 1015
Score = 30.8 bits (68), Expect = 0.73
Identities = 18/38 (47%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Frame = +1
Query: 142 AESYCFWTPKS-SSLIASPKTSPMNSTIKCKKSNSTGS 178
AE C W PKS S+ A+ T+P + IKCK ST S
Sbjct: 232 AELACRWFPKSPSNPTAAATTTPSTAHIKCKWFTSTKS 345
>TC205038 similar to UP|Q94II3 (Q94II3) ERD3 protein, partial (48%)
Length = 889
Score = 30.4 bits (67), Expect = 0.96
Identities = 25/106 (23%), Positives = 40/106 (37%), Gaps = 28/106 (26%)
Frame = -1
Query: 148 WT---PKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKES- 203
WT PKSS +NSTI C ++G+ S+ + W++ SL R + +++
Sbjct: 619 WTRQDPKSSGAKNPVNFKEINSTIFCPWDKASGAMSHIKGA*WEWKSLCRNDTENCRDTT 440
Query: 204 ----LNM--------------------VTPAKKENLKLNSGGNGNV 225
LN+ TPA +N NG +
Sbjct: 439 LQSKLNLSFMVISWCKRKS*NPTI*QVTTPASNSTTSINGSSNGPI 302
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 30.4 bits (67), Expect = 0.96
Identities = 21/57 (36%), Positives = 30/57 (51%)
Frame = -3
Query: 129 TSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSS 185
+SSE + S + S WT SSS +S +SP + S S+ SSS++SSS
Sbjct: 519 SSSESSSESLSSSSGSPPRWTSPSSSSSSSSSSSPPSGASSSSSSLSSPSSSSSSSS 349
Score = 28.9 bits (63), Expect = 2.8
Identities = 25/106 (23%), Positives = 45/106 (41%)
Frame = -3
Query: 150 PKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVTP 209
PKSSS S +S +S+ + S+ + SSS+ S RW S S S S + +
Sbjct: 573 PKSSSSPCSSSSSSSSSSSSSESSSESLSSSSGSPPRWTSPS----SSSSSSSSSSPPSG 406
Query: 210 AKKENLKLNSGGNGNVVGKKIPATEKKTPATVSAMEVFYGRKKETR 255
A + L+S + + ++ +T S+ + + T+
Sbjct: 405 ASSSSSSLSSPSSSS--SSSSSSSSSSASSTTSSTDSYSSSSSSTK 274
>BE609826
Length = 223
Score = 29.6 bits (65), Expect = 1.6
Identities = 15/40 (37%), Positives = 19/40 (47%)
Frame = -3
Query: 150 PKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKF 189
P +S T PM+S I C S S S+ +SS W F
Sbjct: 140 PSASGPECPCHTQPMHSQIACSPVESIWSPSHMTSSNWNF 21
>TC213554 weakly similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1,
partial (18%)
Length = 451
Score = 29.6 bits (65), Expect = 1.6
Identities = 15/40 (37%), Positives = 24/40 (59%)
Frame = -2
Query: 145 YCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSS 184
+ FW+ SSS +S +S +S+ S+S+ SSS +SS
Sbjct: 309 WSFWSSSSSSSSSSSSSSSSSSSTSSSTSSSSTSSSTSSS 190
>TC216975 similar to UP|Q6H974 (Q6H974) SEU1 protein, partial (28%)
Length = 1544
Score = 29.6 bits (65), Expect = 1.6
Identities = 15/37 (40%), Positives = 18/37 (48%)
Frame = +3
Query: 142 AESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGS 178
AE C W PK S + T P + IKCK +T S
Sbjct: 759 AEFIC*WFPKPPSTPTAAATPPTTALIKCKCFTATKS 869
>BQ079935
Length = 407
Score = 29.6 bits (65), Expect = 1.6
Identities = 13/52 (25%), Positives = 27/52 (51%)
Frame = -1
Query: 89 NLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIGNEKEKWTSSEVEDDLDSI 140
N ++G+E K+ G R + ++ ++ + E EKW E+E +++ I
Sbjct: 185 NGMVGEEMEKVVEGSRSGKEVEDMTLVVEGICSNREDEKWEMVEMEMEVEGI 30
>TC207093
Length = 863
Score = 29.3 bits (64), Expect = 2.1
Identities = 16/37 (43%), Positives = 24/37 (64%)
Frame = +3
Query: 149 TPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSS 185
TPK+SS SP T P++S + S++ SS++SSS
Sbjct: 219 TPKTSS-FCSPPTVPISSVVAAAASSTPRHSSSSSSS 326
>TC233023 similar to UP|O48652 (O48652) 6-4 photolyase, partial (39%)
Length = 678
Score = 29.3 bits (64), Expect = 2.1
Identities = 26/87 (29%), Positives = 39/87 (43%), Gaps = 5/87 (5%)
Frame = -2
Query: 139 SIPAESYCFWTPKS-----SSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLL 193
SIPA S F T S S+ S + ++ K + + S GS S +FL L
Sbjct: 296 SIPAASAKFLTLTSRA**YGSVSYSKHSFLTCNSFKHRMTTSAGSPLRIKSLEPRFLRLR 117
Query: 194 RRSKSDGKESLNMVTPAKKENLKLNSG 220
RS ++L PA+++ + NSG
Sbjct: 116 SRSTRHSSKNLMRFRPAREDPGRKNSG 36
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.309 0.126 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,565,909
Number of Sequences: 63676
Number of extensions: 156125
Number of successful extensions: 1307
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 1123
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1220
length of query: 285
length of database: 12,639,632
effective HSP length: 96
effective length of query: 189
effective length of database: 6,526,736
effective search space: 1233553104
effective search space used: 1233553104
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC124951.5


