

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124215.6 - phase: 0 /pseudo
(679 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
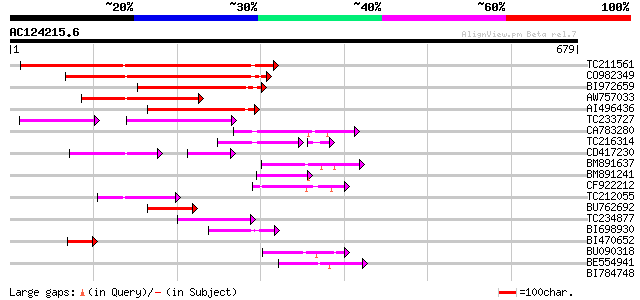
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 286 2e-77
CO982349 209 2e-54
BI972659 142 5e-34
AW757033 134 1e-31
AI496436 114 1e-25
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 67 5e-23
CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polym... 92 8e-19
TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {A... 91 8e-19
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 66 8e-18
BM891637 73 5e-13
BM891241 70 3e-12
CF922212 69 5e-12
TC212055 62 8e-10
BU762692 62 1e-09
TC234877 56 6e-08
BI698930 53 4e-07
BI470652 similar to PIR|T17102|T171 RNA-directed DNA polymerase ... 50 3e-06
BU090318 weakly similar to GP|9743345|gb| F21J9.30 {Arabidopsis ... 47 4e-05
BE554941 46 6e-05
BI784748 39 0.010
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 286 bits (733), Expect = 2e-77
Identities = 144/309 (46%), Positives = 201/309 (64%)
Frame = +1
Query: 13 LDGILVANEVVDDARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFPTLWRKWIKKC 72
L L+ANEV+D+A++ K L+FKVD+EKAYDSV N+L +M +M F W KWI++C
Sbjct: 1 LHSALIANEVIDEAKRSNKSCLVFKVDYEKAYDSVSWNFLMYMMWRMDFSPKWIKWIEEC 180
Query: 73 VGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALVVNNLFNGYKV 132
V +A+ SVLVNGSPT EF RGLRQGDPL+PFLF + AEG + LM V NL+ Y V
Sbjct: 181 VKSASISVLVNGSPTAEFLPQRGLRQGDPLTPFLFNIVAEGLNGLMRRAVEENLYKPYLV 360
Query: 133 GSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFSKSLFVGVNVHG 192
G++ V ++ LQ+ADDTI + + N+ A++ IL F+ +S L++NF+KS F V
Sbjct: 361 GANG-VPISILQYADDTIFFGEAAKENVEAIKVILRSFELVSNLRINFAKSCFGVFGVTD 537
Query: 193 SWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSSWKSKHLSLGGR 252
W EAA L+C+ + PF+YLG+PIG N+ R W +I + +LS WK +H+S GGR
Sbjct: 538 QWKQEAANYLHCSLLAFPFIYLGIPIGANSRRCHMWDSIIRKCERKLSKWK*RHISFGGR 717
Query: 253 LVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGSADHRKIIWVDWNSV 312
+ L+KSVL+S+P+Y SFF+ P +V + I F WGGG D +KI W+ W +
Sbjct: 718 VTLIKSVLTSIPIYFFSFFRVPQSVVDKLVKI*RTFL----WGGG-PDQKKISWIRWEKL 882
Query: 313 CRSQEVGGL 321
C +E GG+
Sbjct: 883 CLPKERGGI 909
>CO982349
Length = 795
Score = 209 bits (533), Expect = 2e-54
Identities = 108/246 (43%), Positives = 154/246 (61%)
Frame = +3
Query: 68 WIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALVVNNLF 127
WI+ C+ +A+ S+LVN SP EFS RGLRQGDPL+P LF + AEG LM V F
Sbjct: 69 WIRGCLHSASVSILVNESPINEFSPQRGLRQGDPLAPLLFNIVAEGLTGLMREAVDRKRF 248
Query: 128 NGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFSKSLFVG 187
N + VG ++ V+ LQ+ADDTI + + N++ ++ IL F+ SGLK+NF++S F
Sbjct: 249 NSFLVGKNKEP-VSILQYADDTIFFEEATMENMKVIKTILRCFELASGLKINFAESRFGA 425
Query: 188 VNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSSWKSKHL 247
+ W EAA LNC+ S+PF YLG+PI N P+I + ++L+ WK +H+
Sbjct: 426 IWKPDHWCKEAAEFLNCSMLSMPFSYLGIPIEANPRCREI*DPIIRKCETKLARWKQRHI 605
Query: 248 SLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGSADHRKIIWV 307
SLGGR+ L+ ++L++LP+Y SFF+APS +V + +I F WGGGS R+I WV
Sbjct: 606 SLGGRVTLINAILTALPIYFFSFFRAPSKVVQKLVNIQRKFL----WGGGSX-QRRIAWV 770
Query: 308 DWNSVC 313
W +VC
Sbjct: 771 KWETVC 788
>BI972659
Length = 453
Score = 142 bits (358), Expect = 5e-34
Identities = 71/154 (46%), Positives = 100/154 (64%)
Frame = +1
Query: 154 DKSWANIRALRAILLLFQELSGLKVNFSKSLFVGVNVHGSWLAEAATVLNCNAGSIPFLY 213
+ SW NI L+++L F+ +SGL++N++KS F V +W +AA +LNC IPF Y
Sbjct: 7 EASWDNIIVLKSMLRGFEMVSGLRINYAKSQFGIVGFQPNWAHDAAQLLNCRQLDIPFHY 186
Query: 214 LGLPIGGNASRMVFWKPLINRINSRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKA 273
LG+PI AS + W+PLIN+ ++LS W K+LS+GGR+ L+KSVL++LP+Y LSFFK
Sbjct: 187 LGMPIAVKASSRMVWEPLINKFKAKLSKWNQKYLSMGGRVTLIKSVLNALPIYLLSFFKI 366
Query: 274 PSGIVSSIESILNNFFFWGGWGGGSADHRKIIWV 307
P IV + S L F W GG+ H +I WV
Sbjct: 367 PQRIVDKLVS-LQRTFMW----GGNQHHNRISWV 453
>AW757033
Length = 441
Score = 134 bits (337), Expect = 1e-31
Identities = 70/146 (47%), Positives = 93/146 (62%)
Frame = -2
Query: 87 TYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALVVNNLFNGYKVGSHEMVGVTHLQFA 146
T EFS HRGLRQGDPL+P LF +AAEG LM V N + + VG ++ V+ LQ+A
Sbjct: 440 TREFSPHRGLRQGDPLAPLLFNIAAEGLTGLMREAVARNHYKSFAVGKYKKP-VSILQYA 264
Query: 147 DDTIILCDKSWANIRALRAILLLFQELSGLKVNFSKSLFVGVNVHGSWLAEAATVLNCNA 206
DDTI + + N+R ++ IL F+ SGLK+NF+KS F ++V W EAA +NC+
Sbjct: 263 DDTIFFGEATMENVRVIKTILRGFELASGLKINFAKSRFGAISVPD*WRKEAAEFMNCSL 84
Query: 207 GSIPFLYLGLPIGGNASRMVFWKPLI 232
S+PF YLG+PIG N R W P+I
Sbjct: 83 LSLPFSYLGIPIGANPRRRETWDPII 6
>AI496436
Length = 414
Score = 114 bits (285), Expect = 1e-25
Identities = 57/134 (42%), Positives = 84/134 (62%)
Frame = +1
Query: 166 ILLLFQELSGLKVNFSKSLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRM 225
IL F+ SGLK+NF+KS F + W EAA LN + S+PF+YLG+PIG N+
Sbjct: 4 ILRCFELSSGLKINFAKSQFGTICKSDIWRKEAADYLNSSVLSMPFVYLGIPIGANSRHS 183
Query: 226 VFWKPLINRINSRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESIL 285
W+P++ + +L+SWK K++S GGR+ L+ SVL++LP+Y LSFF+ P +V +
Sbjct: 184 DVWEPVVRKCERKLASWKQKYVSFGGRVTLINSVLTALPIYLLSFFRIPDKVVDKAD--Y 357
Query: 286 NNFFFWGGWGGGSA 299
N+ + GW G A
Sbjct: 358 NSA*IFMGWRFGIA 399
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 66.6 bits (161), Expect(2) = 5e-23
Identities = 38/97 (39%), Positives = 57/97 (58%), Gaps = 1/97 (1%)
Frame = -3
Query: 12 ILDGILVANEVVDDARKRK-KDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFPTLWRKWIK 70
I D V +EV++ K+ L K+D KA+D++D ++L V+ K G+ + WI+
Sbjct: 834 IKDCTCVTSEVINMLDKKVFGGNLALKIDIVKAFDTLDRDFLISVLNKFGYSHTFCNWIR 655
Query: 71 KCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLF 107
+ +A S+ VNG P FS RG+RQGDPLSP L+
Sbjct: 654 VILLSAKLSISVNGEPVGFFSCQRGVRQGDPLSPSLY 544
Score = 60.1 bits (144), Expect(2) = 5e-23
Identities = 38/132 (28%), Positives = 68/132 (50%), Gaps = 1/132 (0%)
Frame = -2
Query: 141 THLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFSKS-LFVGVNVHGSWLAEAA 199
+H+ ADD +I C + N+ L L+++ G ++ KS +F+G ++ G L +
Sbjct: 451 SHVMHADDIMIFCKGTSRNVHNLINFFQLYKDSFGQVISKHKSKIFIG-SIPGQRLRVIS 275
Query: 200 TVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSSWKSKHLSLGGRLVLLKSV 259
+L IPF Y G+PI + ++I +L+SWK LS+ GR+ L+ SV
Sbjct: 274 DLLQYFHSDIPFNYFGVPIFKGKPTRRHLYCVADKIKCKLASWKGSILSIMGRIQLVNSV 95
Query: 260 LSSLPVYALSFF 271
+ + +Y+ S +
Sbjct: 94 IHGMLLYSFSVY 59
>CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polymerase
homolog T24M8.8 - Arabidopsis thaliana, partial (4%)
Length = 456
Score = 92.0 bits (227), Expect = 8e-19
Identities = 53/158 (33%), Positives = 81/158 (50%), Gaps = 8/158 (5%)
Frame = +2
Query: 269 SFFKAPSGIVSSIESILNNFFFWGGWGGGSADHRKIIWVDWNSVCRSQEVGGLEVRRIKE 328
SFF P I++ +ES+ F + GG AD RKI WV+W +VC + GGL ++ ++
Sbjct: 5 SFFSLP*KIIAKLESLQRRFLW-----GGEADSRKIAWVNWKTVCLPKAKGGLGIKDLRT 169
Query: 329 FNLALLGKWCWRVLTERDSLWFRVLAVR----NRVEGGHLCSGGRNESMWWRNIV---AL 381
FN LLGKW W + + W +VL + +E G S G +S WW++++ L
Sbjct: 170 FNTTLLGKWRWDLFYIQQEPWAKVLQSKYGGWRALEEG---SSGSKDSAWWKDLIKTQQL 340
Query: 382 HSEGWFQNHVSRSLGDGSSVLFWTDV*VG-ELSLRDRF 418
+ +G G + FW D+ +LSLRD+F
Sbjct: 341 QRNIPLKRETIWKVGGGDRIKFWEDLWTNTDLSLRDKF 454
>TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {Arabidopsis
thaliana;} , partial (18%)
Length = 759
Score = 91.3 bits (225), Expect(2) = 8e-19
Identities = 41/104 (39%), Positives = 62/104 (59%)
Frame = +2
Query: 249 LGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGSADHRKIIWVD 308
+GGR+ L+KSVLS+LP+ LSFFK P IV + ++ F + GG+ H +I WV
Sbjct: 2 MGGRVTLIKSVLSALPIXXLSFFKIPQRIVDKLVTLQRQFLW-----GGTQHHNRIPWVK 166
Query: 309 WNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRV 352
W +C + GGL ++ + FN AL G+W W + + + LW R+
Sbjct: 167 WADICNPKIDGGLGIKDLSNFNAALRGRWIWGLASNHNQLWARL 298
Score = 21.2 bits (43), Expect(2) = 8e-19
Identities = 10/32 (31%), Positives = 17/32 (52%)
Frame = +3
Query: 357 NRVEGGHLCSGGRNESMWWRNIVALHSEGWFQ 388
+RV+G H S WW+++ L+ + FQ
Sbjct: 306 SRVKGWH--------SSWWKDLKKLYHQPDFQ 377
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like
protein {Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 66.2 bits (160), Expect(2) = 8e-18
Identities = 37/112 (33%), Positives = 60/112 (53%)
Frame = -3
Query: 72 CVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALVVNNLFNGYK 131
C+ T T S+L NG FS + G+RQ DP++P+LF+L E L+ ++ L +
Sbjct: 668 CMSTTTMSLLWNGEVLDHFSPNIGVRQRDPIAPYLFVLCLERLSHLINLVMNKKL*RSIQ 489
Query: 132 VGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFSKS 183
V ++ ++HL F DD I+ + + + + L LF + SG KVN K+
Sbjct: 488 VNRGGLL-ISHLAFVDDLILFVEANMDQVDVINLTLELFSDSSGAKVNVDKT 336
Score = 42.7 bits (99), Expect(2) = 8e-18
Identities = 23/58 (39%), Positives = 34/58 (57%)
Frame = -2
Query: 213 YLGLPIGGNASRMVFWKPLINRINSRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSF 270
YLG+PI + +I+++N R S WKS LS+ GRL L K V+ +LP + + F
Sbjct: 243 YLGVPIFHKNPTSHTFDFIIDKVNKRFSRWKSNLLSMVGRLTLTK*VV*ALPSHIMQF 70
>BM891637
Length = 427
Score = 72.8 bits (177), Expect = 5e-13
Identities = 44/131 (33%), Positives = 65/131 (49%), Gaps = 7/131 (5%)
Frame = -1
Query: 302 RKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVRNRVEG 361
+KI W+ W C S +VGGL ++ IK N ALL KW W + + LW R+L ++ +G
Sbjct: 418 KKIAWISWRQCCASGDVGGLGIKDIKILNNALLIKWKWLMFHQPHQLWNRILI--SKYKG 245
Query: 362 GHLCSGGRNE---SMWWRNIVALHSEGWF---QNHVSRSLGDGSSVLFWTDV*VGE-LSL 414
G + S WW ++ A++ N +G G +LFW D V + L
Sbjct: 244 WRGLDQGPQKYYFSPWWADLRAINQHQSMIAASNQFCWKVGRGDQILFWEDSWVDDGTPL 65
Query: 415 RDRFSRLYDLS 425
+D+F LY LS
Sbjct: 64 KDQFPELYRLS 32
>BM891241
Length = 407
Score = 70.1 bits (170), Expect = 3e-12
Identities = 31/76 (40%), Positives = 45/76 (58%), Gaps = 9/76 (11%)
Frame = -2
Query: 296 GGSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAV 355
GG DH+KI WV W VC + GGL ++ I +FN+ALLGKW W + +++ LW R++
Sbjct: 406 GGDHDHKKIPWVKWEVVCLPKTKGGLGIKDISKFNIALLGKWIWALASDQQQLWARIINS 227
Query: 356 R---------NRVEGG 362
+ N V+GG
Sbjct: 226 KYGGWSDFQYNSVQGG 179
>CF922212
Length = 445
Score = 69.3 bits (168), Expect = 5e-12
Identities = 47/126 (37%), Positives = 63/126 (49%), Gaps = 10/126 (7%)
Frame = -2
Query: 291 WGGWGGGSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWF 350
W WGGG +KI WV+W+SVC +E GL R +++FN ALLGK W + + L
Sbjct: 399 WFLWGGGLGQ-KKIAWVNWDSVCLPKEKEGLGNRDLRKFNYALLGKRRWNLFHHQGELGA 223
Query: 351 RVL-----AVRNRVEGGHLCSGGRNESMWWRNIVAL-HSE---GWFQNHVSRS-LGDGSS 400
RVL RN E + ++ES WW+ I + HS WF+ LG +
Sbjct: 222 RVLDSKYKRWRNLDEERRV----KSESFWWQEISFITHSTEDGSWFEKRTKTGRLGCRAK 55
Query: 401 VLFWTD 406
V FW D
Sbjct: 54 VRFWED 37
>TC212055
Length = 776
Score = 62.0 bits (149), Expect = 8e-10
Identities = 35/100 (35%), Positives = 57/100 (57%), Gaps = 1/100 (1%)
Frame = +1
Query: 106 LFLLAAEGFHVLMEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRA 165
LF + AEG LM + +L++ VG ++ + V LQ+ADDTI L + + N+ +++
Sbjct: 1 LFNIVAEGLTGLMREALDKSLYSSLMVGKNK-IPVNILQYADDTIFLGEATMQNVMTIKS 177
Query: 166 ILLLFQELSGLKVNFSKSLFVGVNVHGSWLAEAA-TVLNC 204
+L +F+ SGLK++F+KS F + W EA NC
Sbjct: 178 MLRVFELASGLKIHFAKSSFGAIGKPDHWQKEAGLNTFNC 297
>BU762692
Length = 423
Score = 61.6 bits (148), Expect = 1e-09
Identities = 29/59 (49%), Positives = 39/59 (65%)
Frame = -3
Query: 166 ILLLFQELSGLKVNFSKSLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASR 224
IL F+ +SGLK+NF+KS F + W EAA+ LNC+ +IPF+Y G+PIG N R
Sbjct: 181 ILRTFELVSGLKINFAKSCFGAFGMTDRWKTEAASCLNCSLLAIPFVYQGIPIGANPRR 5
>TC234877
Length = 534
Score = 55.8 bits (133), Expect = 6e-08
Identities = 26/94 (27%), Positives = 51/94 (53%)
Frame = +1
Query: 201 VLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSSWKSKHLSLGGRLVLLKSVL 260
+L + GS+PF+YL +P+ R V +P+ +RI + L SWK LS G + L+ S++
Sbjct: 13 ILGFSVGSLPFIYLSVPLFQEKPRPVHLRPIADRIKAELQSWKGNLLSFMGCVQLIISII 192
Query: 261 SSLPVYALSFFKAPSGIVSSIESILNNFFFWGGW 294
+ + + + P ++ +++ NF + G+
Sbjct: 193 DGMLL*SFHLYSWPKALLIEMQTWTRNFIWTRGY 294
>BI698930
Length = 384
Score = 53.1 bits (126), Expect = 4e-07
Identities = 34/87 (39%), Positives = 47/87 (53%), Gaps = 2/87 (2%)
Frame = +3
Query: 239 LSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGS 298
L+ WK + LS GR L+ S+LSSLP+++ SFFK P + SI NF G W
Sbjct: 81 LALWKHEVLSFAGRTCLINSMLSSLPLFSFSFFKVPLNVGMRFISI*RNFV--GEW---- 242
Query: 299 ADHRKIIWVDWNS--VCRSQEVGGLEV 323
+ I WV+W+ V R E GG ++
Sbjct: 243 RES*MIPWVNWDKV*VKRVCEFGGKQI 323
>BI470652 similar to PIR|T17102|T171 RNA-directed DNA polymerase (EC
2.7.7.49) (clone AmLi2) - garden snapdragon (fragment),
partial (22%)
Length = 111
Score = 50.4 bits (119), Expect = 3e-06
Identities = 22/36 (61%), Positives = 27/36 (74%)
Frame = +1
Query: 70 KKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPF 105
K+C+ +A+ S+LVNGSPT EF RGLRQG P PF
Sbjct: 1 KECLSSASISILVNGSPTEEFKPXRGLRQGGPFGPF 108
>BU090318 weakly similar to GP|9743345|gb| F21J9.30 {Arabidopsis thaliana},
partial (1%)
Length = 422
Score = 46.6 bits (109), Expect = 4e-05
Identities = 32/109 (29%), Positives = 55/109 (50%), Gaps = 4/109 (3%)
Frame = +3
Query: 303 KIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVRNRVEGG 362
K+ W W ++C ++ GGL +R + N +LL K + T+ + L RV VR++ + G
Sbjct: 3 KVHWTSWKALCSPKKSGGLGLRLLSHMNKSLLMKST*NMCTKPNDL*VRV--VRSKYK*G 176
Query: 363 HLC----SGGRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTDV 407
+ + +W+ IV + N ++R +G+G SV FW DV
Sbjct: 177 KDILP*IDPKKTGTNFWQGIVHCWEKITHDN-IARKIGNGRSVHFWKDV 320
>BE554941
Length = 383
Score = 45.8 bits (107), Expect = 6e-05
Identities = 30/111 (27%), Positives = 51/111 (45%), Gaps = 5/111 (4%)
Frame = +1
Query: 323 VRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVR-NRVEGGHLCSGGRNESMWWRNIVAL 381
++ + FN++LL K L E ++W +L ++ + S R S WWR+I+ +
Sbjct: 10 IKNLSIFNISLLVKRR*HFLVETKAIWAPILKFHYGSLDLSNTGSASRT-SQWWRDILRI 186
Query: 382 ----HSEGWFQNHVSRSLGDGSSVLFWTDV*VGELSLRDRFSRLYDLSVLK 428
+ GW + R +GD LFW + E L +RL L++ K
Sbjct: 187 GNCQYQPGWLTSTFCRKIGDRKDTLFWRHRCLNETPLMQSHARLXSLAMDK 339
>BI784748
Length = 259
Score = 38.5 bits (88), Expect = 0.010
Identities = 15/33 (45%), Positives = 27/33 (81%)
Frame = +2
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRKKDLLL 35
QSAFI+GR +L +++ANEV+D+A++ +K ++
Sbjct: 14 QSAFIQGRHMLHSMMIANEVIDEAKRGQKPSIM 112
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.340 0.151 0.516
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,304,431
Number of Sequences: 63676
Number of extensions: 583751
Number of successful extensions: 5016
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 4903
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4999
length of query: 679
length of database: 12,639,632
effective HSP length: 104
effective length of query: 575
effective length of database: 6,017,328
effective search space: 3459963600
effective search space used: 3459963600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC124215.6


