

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124215.2 + phase: 0
(358 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
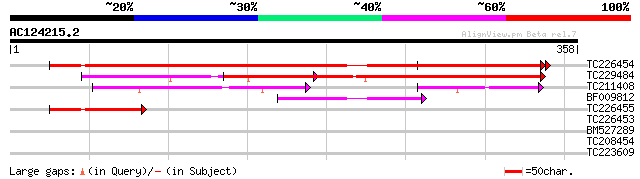
Score E
Sequences producing significant alignments: (bits) Value
TC226454 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine cytid... 416 e-117
TC229484 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine cytid... 359 1e-99
TC211408 similar to UP|Q8L4J8 (Q8L4J8) CTP:phosphorylcholine cyt... 110 8e-25
BF009812 80 1e-15
TC226455 homologue to UP|Q84JV7 (Q84JV7) CTP:ethanolamine cytidy... 70 1e-12
TC226453 weakly similar to UP|Q84JV7 (Q84JV7) CTP:ethanolamine c... 39 0.005
BM527289 28 4.9
TC208454 similar to UP|Q94BU7 (Q94BU7) AT3g10970/F9F8_21, partia... 28 8.4
TC223609 similar to UP|Q9SIA7 (Q9SIA7) Legumin-like protein, par... 28 8.4
>TC226454 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine
cytidylyltransferase, partial (71%)
Length = 1889
Score = 416 bits (1069), Expect = e-117
Identities = 202/313 (64%), Positives = 249/313 (79%)
Frame = +1
Query: 26 VGVSVLGFYSSSGPFGLWGKNGRRRNKKPIRVYMDGCFDMMHYGHCNALRQARALGDQLI 85
+GV L S G++ K ++ K+ IRVYMDGCFD+MHYGH NALRQA+ALGD+L+
Sbjct: 340 IGVPYLSLPCSWSNLGIFHK--KKSGKRRIRVYMDGCFDLMHYGHANALRQAKALGDELV 513
Query: 86 VGVVSDDEIIANKGPPVTPLHERLIMVNAVKWVDEVIPEAPYAITEEFMKKLFDEYNIDY 145
VG+VSD+EI+ANKGPPV + ERL +V+ +KWVDEVI +APYAITE+F+ +LF EY IDY
Sbjct: 514 VGLVSDEEIVANKGPPVLSMEERLALVSGLKWVDEVITDAPYAITEQFLNRLFHEYKIDY 693
Query: 146 IIHGDDPCVLPDGTDAYAHAKKAGRYKQIKRTEGVSSTDIVGRMLLCVRERNIADTHNHS 205
+IHGDDPC+LPDGTDAYA AKKAGRYKQIKRTEGVSSTDIVGR+L +R++ + HN +
Sbjct: 694 VIHGDDPCLLPDGTDAYAAAKKAGRYKQIKRTEGVSSTDIVGRILSSLRDQKNCEDHNGT 873
Query: 206 SLQRQFSNGRGQQKFEDGGVVASGTRVSHFLPTSRRIVQFSNGRSPGPDARIVYIDGAFD 265
++ Q N + + ++ FLPTSRRIVQFSNG+ PGP++RIVYIDGAFD
Sbjct: 874 EVKPQEEN------------QSQASHIAQFLPTSRRIVQFSNGKGPGPNSRIVYIDGAFD 1017
Query: 266 LFHAGHVEILRLARDRGDFLLVGIHTDQTVSATRGLHRPIMSLHERSLSVLACRYVDEVI 325
LFHAGHVEIL+ AR+ GDFLLVGIH+D+TVS RG H PIM LHERSLSVLACRYVDEVI
Sbjct: 1018LFHAGHVEILKRARELGDFLLVGIHSDETVSEHRGNHYPIMHLHERSLSVLACRYVDEVI 1197
Query: 326 IGAPWEVSKDMVS 338
IG+PWE++ DM++
Sbjct: 1198IGSPWEITNDMIT 1236
Score = 70.5 bits (171), Expect = 1e-12
Identities = 34/84 (40%), Positives = 54/84 (63%)
Frame = +1
Query: 258 VYIDGAFDLFHAGHVEILRLARDRGDFLLVGIHTDQTVSATRGLHRPIMSLHERSLSVLA 317
VY+DG FDL H GH LR A+ GD L+VG+ +D+ + A +G P++S+ ER V
Sbjct: 427 VYMDGCFDLMHYGHANALRQAKALGDELVVGLVSDEEIVANKG--PPVLSMEERLALVSG 600
Query: 318 CRYVDEVIIGAPWEVSKDMVSLIF 341
++VDEVI AP+ +++ ++ +F
Sbjct: 601 LKWVDEVITDAPYAITEQFLNRLF 672
>TC229484 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine
cytidylyltransferase, partial (61%)
Length = 947
Score = 359 bits (921), Expect = 1e-99
Identities = 180/206 (87%), Positives = 189/206 (91%), Gaps = 3/206 (1%)
Frame = +2
Query: 136 KLFDEYNIDYIIHGDDPCVLPDGTDAYAHAKKAGRYKQIKRTEGVSSTDIVGRMLLCVRE 195
KLFDEY ID+IIHGDDPCVLPDGTDAYAHAKKAGRYKQIKRTEGVSSTDIVGRMLLCVRE
Sbjct: 2 KLFDEYKIDFIIHGDDPCVLPDGTDAYAHAKKAGRYKQIKRTEGVSSTDIVGRMLLCVRE 181
Query: 196 RNIAD-THNHSSLQRQFSNGRGQQKFEDG--GVVASGTRVSHFLPTSRRIVQFSNGRSPG 252
R+IA+ HNHSSLQRQFSNG KFE G ASGTR+SHFLPTSRRIVQFSNGR PG
Sbjct: 182 RSIAEKNHNHSSLQRQFSNGHSP-KFEAGVSAATASGTRISHFLPTSRRIVQFSNGRGPG 358
Query: 253 PDARIVYIDGAFDLFHAGHVEILRLARDRGDFLLVGIHTDQTVSATRGLHRPIMSLHERS 312
PD+ IVYIDGAFDLFHAGHVEILRLARD GDFLLVGIHTDQTVSATRG HRPIM+LHERS
Sbjct: 359 PDSHIVYIDGAFDLFHAGHVEILRLARDLGDFLLVGIHTDQTVSATRGSHRPIMNLHERS 538
Query: 313 LSVLACRYVDEVIIGAPWEVSKDMVS 338
LSVLACRYVDEVIIGAPWE+SKDM++
Sbjct: 539 LSVLACRYVDEVIIGAPWEISKDMLT 616
Score = 85.1 bits (209), Expect = 4e-17
Identities = 54/155 (34%), Positives = 82/155 (52%), Gaps = 5/155 (3%)
Frame = +2
Query: 46 NGRRRNKKPIRVYMDGCFDMMHYGHCNALRQARALGDQLIVGVVSDDEIIANKGP--PVT 103
NGR VY+DG FD+ H GH LR AR LGD L+VG+ +D + A +G P+
Sbjct: 341 NGRGPGPDSHIVYIDGAFDLFHAGHVEILRLARDLGDFLLVGIHTDQTVSATRGSHRPIM 520
Query: 104 PLHERLIMVNAVKWVDEVIPEAPYAITEEFMKKLFDEYNIDYIIHG---DDPCVLPDGTD 160
LHER + V A ++VDEVI AP+ E K + +NI ++HG + + +
Sbjct: 521 NLHERSLSVLACRYVDEVIIGAPW----EISKDMLTTFNISLVVHGTIAESNDFKKEECN 688
Query: 161 AYAHAKKAGRYKQIKRTEGVSSTDIVGRMLLCVRE 195
YA G +K ++ +++T I+ R ++C E
Sbjct: 689 PYAVPISMGIFKVLESPLDITTTTII-RRIVCNHE 790
>TC211408 similar to UP|Q8L4J8 (Q8L4J8) CTP:phosphorylcholine
cytidylyltransferase , partial (83%)
Length = 870
Score = 110 bits (276), Expect = 8e-25
Identities = 57/142 (40%), Positives = 84/142 (59%), Gaps = 4/142 (2%)
Frame = +2
Query: 53 KPIRVYMDGCFDMMHYGHCNALRQARAL--GDQLIVGVVSDDEIIANKGPPVTPLHERLI 110
+P+RVY DG +D+ H+GH +L QA+ L+VG +D KG V ER
Sbjct: 92 RPVRVYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCNDQVTHKYKGKTVMTEAERYE 271
Query: 111 MVNAVKWVDEVIPEAPYAITEEFMKKLFDEYNIDYIIHGDDPCVLPDG--TDAYAHAKKA 168
+ KWVDEVIP+AP+ I +EF+ D++NIDY+ H P +G D Y K
Sbjct: 272 SLRHCKWVDEVIPDAPWVINQEFL----DKHNIDYVAHDSLPYADANGAANDVYEFVKSV 439
Query: 169 GRYKQIKRTEGVSSTDIVGRML 190
GR+K+ KRTEG+S++D++ R++
Sbjct: 440 GRFKETKRTEGISTSDVIMRIV 505
Score = 58.5 bits (140), Expect = 4e-09
Identities = 32/82 (39%), Positives = 47/82 (57%), Gaps = 2/82 (2%)
Frame = +2
Query: 258 VYIDGAFDLFHAGHVEILRLARDR--GDFLLVGIHTDQTVSATRGLHRPIMSLHERSLSV 315
VY DG +DLFH GH L A+ +LLVG DQ +G + +M+ ER S+
Sbjct: 104 VYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCNDQVTHKYKG--KTVMTEAERYESL 277
Query: 316 LACRYVDEVIIGAPWEVSKDMV 337
C++VDEVI APW ++++ +
Sbjct: 278 RHCKWVDEVIPDAPWVINQEFL 343
>BF009812
Length = 247
Score = 80.1 bits (196), Expect = 1e-15
Identities = 43/94 (45%), Positives = 57/94 (59%)
Frame = +2
Query: 170 RYKQIKRTEGVSSTDIVGRMLLCVRERNIADTHNHSSLQRQFSNGRGQQKFEDGGVVASG 229
RYKQIKRTEGVSSTDI GR+L +R++ D HN + ++ Q N +
Sbjct: 2 RYKQIKRTEGVSSTDIEGRILSSLRDQKSCDDHNGTEVKPQEEN------------QSQA 145
Query: 230 TRVSHFLPTSRRIVQFSNGRSPGPDARIVYIDGA 263
++ F+PT RRIVQ NG+ P P+ RI Y+DGA
Sbjct: 146 WHIAQFVPTCRRIVQL*NGKGPDPNLRIEYMDGA 247
>TC226455 homologue to UP|Q84JV7 (Q84JV7) CTP:ethanolamine
cytidylyltransferase (CTP-phosphoethanolamine
cytidylyltransferase), partial (7%)
Length = 593
Score = 70.1 bits (170), Expect = 1e-12
Identities = 33/61 (54%), Positives = 44/61 (72%)
Frame = +1
Query: 26 VGVSVLGFYSSSGPFGLWGKNGRRRNKKPIRVYMDGCFDMMHYGHCNALRQARALGDQLI 85
+GV L S G++ + ++ K+ IRVYMDGCFD+MHYGH NALRQA+ALGD+L+
Sbjct: 415 IGVPYLSLPCSWSNLGIFHQ--KKSGKRRIRVYMDGCFDLMHYGHANALRQAKALGDELV 588
Query: 86 V 86
V
Sbjct: 589 V 591
Score = 37.7 bits (86), Expect = 0.008
Identities = 17/30 (56%), Positives = 20/30 (66%)
Frame = +1
Query: 258 VYIDGAFDLFHAGHVEILRLARDRGDFLLV 287
VY+DG FDL H GH LR A+ GD L+V
Sbjct: 502 VYMDGCFDLMHYGHANALRQAKALGDELVV 591
>TC226453 weakly similar to UP|Q84JV7 (Q84JV7) CTP:ethanolamine
cytidylyltransferase (CTP-phosphoethanolamine
cytidylyltransferase), partial (11%)
Length = 620
Score = 38.5 bits (88), Expect = 0.005
Identities = 14/20 (70%), Positives = 19/20 (95%)
Frame = +2
Query: 319 RYVDEVIIGAPWEVSKDMVS 338
RYVDEVIIG+PWE++ DM++
Sbjct: 2 RYVDEVIIGSPWEITNDMIT 61
>BM527289
Length = 422
Score = 28.5 bits (62), Expect = 4.9
Identities = 13/50 (26%), Positives = 26/50 (52%)
Frame = +3
Query: 301 LHRPIMSLHERSLSVLACRYVDEVIIGAPWEVSKDMVSLIFACLFFMLFV 350
L++PIM +H R LS+ CR +D W + + ++ + C+ + +
Sbjct: 282 LNQPIMEVH*RELSIHTCRMLD-----IRWGLKQWAMTRVLVCICSQMLI 416
>TC208454 similar to UP|Q94BU7 (Q94BU7) AT3g10970/F9F8_21, partial (32%)
Length = 856
Score = 27.7 bits (60), Expect = 8.4
Identities = 17/51 (33%), Positives = 23/51 (44%)
Frame = -1
Query: 43 WGKNGRRRNKKPIRVYMDGCFDMMHYGHCNALRQARALGDQLIVGVVSDDE 93
W G+RR K IRVY C + YG C + G+ G+ DD+
Sbjct: 274 WEVKGKRRGLKAIRVYWRNCM-RIGYGICES-------GEVHRFGINDDDD 146
>TC223609 similar to UP|Q9SIA7 (Q9SIA7) Legumin-like protein, partial (37%)
Length = 717
Score = 27.7 bits (60), Expect = 8.4
Identities = 24/80 (30%), Positives = 36/80 (45%), Gaps = 2/80 (2%)
Frame = +1
Query: 242 IVQFSNGRSPGPDARIVYIDGAFDLFHAGHVEILRLARDRGDFLLVGIHTDQTVSATRGL 301
I+ S+G P I+Y+ ++ G + LR + L + I S RGL
Sbjct: 424 ILMDSSGSLSSPPL-ILYLPTWLEVLPCGRLYHLRFCK----LLSM*IQKLSNCSVQRGL 588
Query: 302 HRPIMSLHERSLSV--LACR 319
P SLH+ LS+ LAC+
Sbjct: 589 PMPFSSLHQTRLSIPLLACK 648
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.141 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,754,319
Number of Sequences: 63676
Number of extensions: 215475
Number of successful extensions: 1034
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 1012
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1023
length of query: 358
length of database: 12,639,632
effective HSP length: 98
effective length of query: 260
effective length of database: 6,399,384
effective search space: 1663839840
effective search space used: 1663839840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC124215.2


