

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123898.9 + phase: 0 /pseudo
(194 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
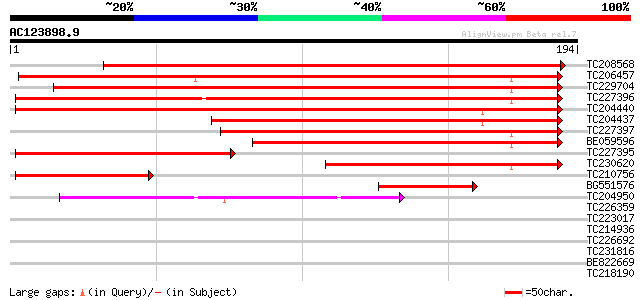
Score E
Sequences producing significant alignments: (bits) Value
TC208568 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {A... 270 3e-73
TC206457 weakly similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannit... 184 2e-47
TC229704 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehy... 175 1e-44
TC227396 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehy... 174 2e-44
TC204440 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehy... 163 5e-41
TC204437 homologue to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol de... 115 2e-26
TC227397 similar to UP|MTD1_STYHU (Q43137) Probable mannitol deh... 97 4e-21
BE059596 89 1e-18
TC227395 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehy... 89 1e-18
TC230620 similar to UP|Q9ATW1 (Q9ATW1) Cinnamyl alcohol dehydrog... 70 6e-13
TC210756 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehy... 49 2e-06
BG551576 homologue to SP|P30359|CAD4_ Cinnamyl-alcohol dehydroge... 44 6e-05
TC204950 similar to UP|Q945P3 (Q945P3) At1g23740/F5O8_27, partia... 42 2e-04
TC226359 similar to UP|O23939 (O23939) Ripening-induced protein ... 37 0.007
TC223017 similar to PIR|T49047|T49047 quinone reductase-like pro... 36 0.013
TC214936 UP|O82478 (O82478) Alcohol dehydrogenase Adh-1 (Fragme... 34 0.049
TC226692 similar to UP|Q9AYU1 (Q9AYU1) Quinone-oxidoreductase QR... 33 0.083
TC231816 similar to UP|Q9MBD7 (Q9MBD7) NAD-dependent sorbitol de... 33 0.11
BE822669 weakly similar to GP|17978983|gb At1g79870/F19K16_17 {A... 32 0.14
TC218190 similar to UP|TIPA_PHAVU (P23958) Probable aquaporin TI... 31 0.31
>TC208568 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {Arabidopsis
thaliana;} , partial (52%)
Length = 556
Score = 270 bits (690), Expect = 3e-73
Identities = 134/158 (84%), Positives = 145/158 (90%)
Frame = +1
Query: 33 LIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVVGLGGLGHMAVKFGKAFGLRV 92
+IPKSYPLA+A PLLCAGITVYSPM+RH MNQPGKSLGV+GLGGLGHMAVKFGKAFGL V
Sbjct: 1 MIPKSYPLAAAAPLLCAGITVYSPMVRHKMNQPGKSLGVIGLGGLGHMAVKFGKAFGLSV 180
Query: 93 TVFSTSMSKKEEALSFLGADQFVVSSNQEEMRALAKTLDFIIDTASGDHPFDPYMSLLKI 152
TVFSTS+SKKEEALS LGAD+FVVSSNQEEM ALAK+LDFIIDTASGDH FDPYMSLLK
Sbjct: 181 TVFSTSISKKEEALSLLGADKFVVSSNQEEMTALAKSLDFIIDTASGDHSFDPYMSLLKT 360
Query: 153 SGVLALVGFPSEVKFSPASLNLGSRTVAGSAAGGTKEI 190
GV LVGFPS+VKF PASLN+GS+TVAGS GGTK+I
Sbjct: 361 YGVFVLVGFPSQVKFIPASLNIGSKTVAGSVTGGTKDI 474
>TC206457 weakly similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol
dehydrogenase (NAD-dependent mannitol dehydrogenase) ,
partial (75%)
Length = 1180
Score = 184 bits (467), Expect = 2e-47
Identities = 97/188 (51%), Positives = 129/188 (68%), Gaps = 2/188 (1%)
Frame = +3
Query: 4 YTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNM- 62
+ +N V +DG+IT GGYS V + RY IP++ + +A PLLCAGITV++P+ H++
Sbjct: 180 FVYNGVFWDGSITYGGYSQIFVADYRYVVHIPENLAMDAAAPLLCAGITVFNPLKDHDLV 359
Query: 63 NQPGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEE 122
PGK +GVVGLGGLGH+AVKFGKAFG VTV STS SK+ EA LGAD F+VSSN ++
Sbjct: 360 ASPGKKIGVVGLGGLGHIAVKFGKAFGHHVTVISTSPSKEAEAKQRLGADDFIVSSNPKQ 539
Query: 123 MRALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVGFPSEVKFSPA-SLNLGSRTVAG 181
++A +++DFI+DT S +H P + LLK++G L LVG P + PA L G R+V G
Sbjct: 540 LQAARRSIDFILDTVSAEHSLLPILELLKVNGTLFLVGAPDKPLQLPAFPLIFGKRSVKG 719
Query: 182 SAAGGTKE 189
GG KE
Sbjct: 720 GIIGGIKE 743
>TC229704 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , partial (63%)
Length = 816
Score = 175 bits (444), Expect = 1e-44
Identities = 94/175 (53%), Positives = 118/175 (66%), Gaps = 1/175 (0%)
Frame = +2
Query: 16 TKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVVGLG 75
T GGYS S+V +E + IP PL +A PLLCAGITVYSP+ + +++PG +GVVGLG
Sbjct: 2 TYGGYSDSMVADEHFVVRIPDRLPLDAAAPLLCAGITVYSPLRYYGLDKPGLHVGVVGLG 181
Query: 76 GLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRALAKTLDFIID 135
GLGHMAVKF KAFG +VTV STS +KKEEA+ LGAD F++S +Q++M+A TLD IID
Sbjct: 182 GLGHMAVKFAKAFGAKVTVISTSPNKKEEAIQNLGADSFLISRDQDQMQAAMGTLDGIID 361
Query: 136 TASGDHPFDPYMSLLKISGVLALVGFPSEVKFSPA-SLNLGSRTVAGSAAGGTKE 189
T S HP P + LLK G L +VG P + P L G + VAG+ GG E
Sbjct: 362 TVSAVHPLLPLIGLLKSHGKLVMVGAPEKPLELPVFPLLAGRKIVAGTLIGGLME 526
>TC227396 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , partial (70%)
Length = 927
Score = 174 bits (442), Expect = 2e-44
Identities = 92/188 (48%), Positives = 125/188 (65%), Gaps = 1/188 (0%)
Frame = +2
Query: 3 VYTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNM 62
V+T+N YDGT T+GGYS +VV++RY P++ PL + PLLCAGITVYSPM + M
Sbjct: 53 VFTYNSPYYDGTRTQGGYSNIVVVHQRYVLRFPENLPLDAGAPLLCAGITVYSPMKYYGM 232
Query: 63 NQPGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEE 122
+P ++GV GL GLGH+A+K KAFGL+VTV S+S +K+ EA+ LGAD F+VSS+ +
Sbjct: 233 TEPA-NIGVEGLVGLGHVAIKLAKAFGLKVTVISSSPNKQAEAIDRLGADSFLVSSDPAK 409
Query: 123 MRALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVGFPSEVKFSPA-SLNLGSRTVAG 181
M+ T+D+IIDT S H P + LLK++G L VG P++ P L G + + G
Sbjct: 410 MKVALGTMDYIIDTISAVHSLIPLLGLLKLNGKLVTVGLPNKPLELPIFPLVAGRKLIGG 589
Query: 182 SAAGGTKE 189
S GG KE
Sbjct: 590 SNFGGIKE 613
>TC204440 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (97%)
Length = 1481
Score = 163 bits (412), Expect = 5e-41
Identities = 81/188 (43%), Positives = 122/188 (64%), Gaps = 1/188 (0%)
Frame = +3
Query: 3 VYTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNM 62
++++N V DG T+GG++ +++V +++ IP+ PLLCAG+TVYSP++ +
Sbjct: 411 IWSYNDVYVDGKPTQGGFAETMIVEQKFVVKIPEGLAPEQVAPLLCAGVTVYSPLVHFGL 590
Query: 63 NQPGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEE 122
+ G G++GLGG+GHM VK KA G VTV S+S KK+EAL LGADQ++VSS+
Sbjct: 591 KESGLRGGILGLGGVGHMGVKIAKALGHHVTVISSSDKKKQEALEHLGADQYLVSSDATA 770
Query: 123 MRALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVG-FPSEVKFSPASLNLGSRTVAG 181
M+ A +LD+IIDT HP +PY+SLLK+ G L L+G + ++F + LG +++ G
Sbjct: 771 MQEAADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFVSPMVMLGRKSITG 950
Query: 182 SAAGGTKE 189
S G KE
Sbjct: 951 SFIGSMKE 974
>TC204437 homologue to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (49%)
Length = 844
Score = 115 bits (287), Expect = 2e-26
Identities = 60/121 (49%), Positives = 81/121 (66%), Gaps = 1/121 (0%)
Frame = +3
Query: 70 GVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRALAKT 129
G++GLGG+GHM VK KA G VTV S+S KK+EAL LGADQ++VSS+ M+ A +
Sbjct: 21 GILGLGGVGHMGVKIAKALGHHVTVISSSDKKKQEALEHLGADQYLVSSDVTAMQEAADS 200
Query: 130 LDFIIDTASGDHPFDPYMSLLKISGVLALVG-FPSEVKFSPASLNLGSRTVAGSAAGGTK 188
LD+IIDT HP +PY+SLLK+ G L L+G + ++F + LG R++ GS G K
Sbjct: 201 LDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFVSPMVMLGRRSITGSFIGSMK 380
Query: 189 E 189
E
Sbjct: 381 E 383
>TC227397 similar to UP|MTD1_STYHU (Q43137) Probable mannitol dehydrogenase 1
(NAD-dependent mannitol dehydrogenase 1) , partial
(48%)
Length = 742
Score = 97.4 bits (241), Expect = 4e-21
Identities = 55/118 (46%), Positives = 72/118 (60%), Gaps = 1/118 (0%)
Frame = +2
Query: 73 GLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRALAKTLDF 132
GL GL H K KAFGL+VTV S+S K+ EA+ LGAD F+VSS+ +M+A T+D+
Sbjct: 2 GLXGLXHXXXKLAKAFGLKVTVISSSPXKQAEAIDRLGADFFLVSSDPAKMKAALGTMDY 181
Query: 133 IIDTASGDHPFDPYMSLLKISGVLALVGFPSEVKFSPA-SLNLGSRTVAGSAAGGTKE 189
IIDT S H P + LLK++G L VG P++ P L G + + GS GG KE
Sbjct: 182 IIDTISAVHSLIPLLGLLKLNGKLVTVGLPNKPLELPIFPLVAGRKLIGGSNFGGLKE 355
>BE059596
Length = 466
Score = 89.0 bits (219), Expect = 1e-18
Identities = 49/107 (45%), Positives = 66/107 (60%), Gaps = 1/107 (0%)
Frame = +2
Query: 84 FGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRALAKTLDFIIDTASGDHPF 143
FGKAFG VTV ST SK+ EA LGAD F++S N ++++A ++DFI+DT +H
Sbjct: 2 FGKAFGHHVTVISTCPSKEPEAKQRLGADHFILSCNPKQLQAARTSMDFILDTVVAEHSL 181
Query: 144 DPYMSLLKISGVLALVGFPSEVKFSPA-SLNLGSRTVAGSAAGGTKE 189
P + LLK++G LVG P + SPA L G R+V G GG K+
Sbjct: 182 LPILELLKVNGT*FLVGAPDKPLQSPAFPLIFGKRSVKGGIIGGIKK 322
>TC227395 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , partial (52%)
Length = 1002
Score = 89.0 bits (219), Expect = 1e-18
Identities = 41/75 (54%), Positives = 53/75 (70%)
Frame = +1
Query: 3 VYTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNM 62
V+T+N YDGT T+GGYS +VV++RY P++ PL + PLLCAGIT YSPM + M
Sbjct: 394 VFTYNSPYYDGTRTQGGYSNIMVVHQRYVLRFPENLPLDAGAPLLCAGITXYSPMKYYGM 573
Query: 63 NQPGKSLGVVGLGGL 77
+PGK LGV LGG+
Sbjct: 574 TEPGKHLGVARLGGV 618
>TC230620 similar to UP|Q9ATW1 (Q9ATW1) Cinnamyl alcohol dehydrogenase,
partial (37%)
Length = 690
Score = 70.1 bits (170), Expect = 6e-13
Identities = 39/82 (47%), Positives = 51/82 (61%), Gaps = 1/82 (1%)
Frame = +1
Query: 109 LGADQFVVSSNQEEMRALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVGFPSEVKFS 168
+GAD FVVS Q++M+A+ T+D IIDT S HP P + LLK G L +VG P +
Sbjct: 10 IGADSFVVSREQDQMQAVMGTMDGIIDTVSAVHPLVPLIGLLKPHGKLVMVGAPEKPLEL 189
Query: 169 PA-SLNLGSRTVAGSAAGGTKE 189
P SL +G + V GS+ GG KE
Sbjct: 190 PVFSLLMGRKMVGGSSIGGMKE 255
>TC210756 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , partial (45%)
Length = 549
Score = 48.5 bits (114), Expect = 2e-06
Identities = 22/47 (46%), Positives = 30/47 (63%)
Frame = +3
Query: 3 VYTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCA 49
+ T+ +DGTIT GGYS +V +E + IP + PL +A PLLCA
Sbjct: 408 ILTYGVKYFDGTITHGGYSDLMVADEHFVVRIPDNLPLDAAAPLLCA 548
>BG551576 homologue to SP|P30359|CAD4_ Cinnamyl-alcohol dehydrogenase (EC
1.1.1.195) (CAD). [Common tobacco] {Nicotiana tabacum},
partial (14%)
Length = 352
Score = 43.5 bits (101), Expect = 6e-05
Identities = 19/34 (55%), Positives = 25/34 (72%)
Frame = +1
Query: 127 AKTLDFIIDTASGDHPFDPYMSLLKISGVLALVG 160
A +LD+IIDT HP +PY+SLLK+ G L L+G
Sbjct: 25 ADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMG 126
>TC204950 similar to UP|Q945P3 (Q945P3) At1g23740/F5O8_27, partial (80%)
Length = 1560
Score = 41.6 bits (96), Expect = 2e-04
Identities = 34/119 (28%), Positives = 53/119 (43%), Gaps = 1/119 (0%)
Frame = +1
Query: 18 GGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVV-GLGG 76
G + V E+ PK+ A A L A T Y + R + PGKS+ V+ G GG
Sbjct: 589 GSLAEYTAVEEKLLAPKPKNLDFAQAASLPLAIETAYEGLERTGFS-PGKSILVLNGSGG 765
Query: 77 LGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRALAKTLDFIID 135
+G + ++ K V +TS ++ + L LGAD + +E L + D + D
Sbjct: 766 VGSLVIQLAKQVYGASRVAATSSTRNLDLLKSLGAD-LAIDYTKENFEDLPEKFDVVYD 939
>TC226359 similar to UP|O23939 (O23939) Ripening-induced protein (Fragment),
partial (93%)
Length = 1355
Score = 36.6 bits (83), Expect = 0.007
Identities = 33/120 (27%), Positives = 53/120 (43%), Gaps = 1/120 (0%)
Frame = +3
Query: 18 GGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVVG-LGG 76
G + ER P++ A A L T Y + R + GKS+ V+G GG
Sbjct: 465 GSLAEYTAAEERLLAHKPQNLSFAEAASLPLTLETAYEGLERTGFSA-GKSILVLGGAGG 641
Query: 77 LGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRALAKTLDFIIDT 136
+G ++ K V +T+ ++K E L LGAD + + +E L++ D + DT
Sbjct: 642 VGTHVIQLAKHVYGASKVAATASTRKLELLRNLGAD-WPIDYTKENFEDLSEKFDVVYDT 818
>TC223017 similar to PIR|T49047|T49047 quinone reductase-like protein -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(57%)
Length = 780
Score = 35.8 bits (81), Expect = 0.013
Identities = 31/109 (28%), Positives = 51/109 (46%), Gaps = 9/109 (8%)
Frame = +1
Query: 75 GGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRAL-------- 126
GG+G AV+ GKA G V + ++K + L LG D V N+ +++
Sbjct: 49 GGVGLAAVQIGKACGAIVIAVARG-AEKVQLLKSLGVDHVVDLGNENVTQSIKQFLQARK 225
Query: 127 AKTLDFIIDTASGDHPFDPYMSLLKISGVLALVGFPS-EVKFSPASLNL 174
K +D + D G + + LLK + ++GF S E+ PA++ L
Sbjct: 226 LKGIDVLYDPVGGKLTKES-LRLLKWGAHILIIGFASGEIPLIPANIAL 369
>TC214936 UP|O82478 (O82478) Alcohol dehydrogenase Adh-1 (Fragment) ,
complete
Length = 1353
Score = 33.9 bits (76), Expect = 0.049
Identities = 41/178 (23%), Positives = 72/178 (40%), Gaps = 9/178 (5%)
Frame = +3
Query: 20 YSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVVGLGGLGH 79
+S VV+ I + PL L C T + + +PG S+ + GLG +
Sbjct: 477 FSEYTVVHAGCVAKINPAAPLDKVCVLSCGICTGFGATVNVAKPKPGSSVAIFGLGAVAL 656
Query: 80 MAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEE--MRALAKTLDFIID-- 135
A + + G + +S + E G ++FV + ++ + +A+ + +D
Sbjct: 657 AAAEGARVSGASRIIGVDLVSARFEEAKKFGVNEFVNPKDHDKPVQQVIAEMTNGGVDRA 836
Query: 136 ---TASGDHPFDPYMSLLKISGVLALVGFPS-EVKFSPASLN-LGSRTVAGSAAGGTK 188
T S + + G+ LVG PS + F A +N L RT+ G+ G K
Sbjct: 837 VECTGSIQAMVSAFECVHDGWGLAVLVGVPSKDDAFKTAPINFLNERTLKGTFYGNYK 1010
>TC226692 similar to UP|Q9AYU1 (Q9AYU1) Quinone-oxidoreductase QR1
(Fragment), partial (90%)
Length = 1229
Score = 33.1 bits (74), Expect = 0.083
Identities = 37/143 (25%), Positives = 58/143 (39%), Gaps = 12/143 (8%)
Frame = +1
Query: 18 GGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMR------HNMNQPGKSLGV 71
GG++ V +E P A A L AG+T + + Q L
Sbjct: 343 GGFAEFAVASESLTAARPSEVSAAEAAALPIAGLTARDALTQIAGVKLDGTGQLKNILVT 522
Query: 72 VGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRAL----A 127
GG+GH AV+ K VT +T ++ + + LGAD+ V+ E AL
Sbjct: 523 AASGGVGHYAVQLAKLGNTHVT--ATCGARNIDFVKGLGADE-VLDYRTPEGAALKSPSG 693
Query: 128 KTLDFIIDTASGD--HPFDPYMS 148
+ D +I+ +G FDP ++
Sbjct: 694 RKYDAVINCTTGISWSTFDPNLT 762
>TC231816 similar to UP|Q9MBD7 (Q9MBD7) NAD-dependent sorbitol dehydrogenase,
partial (59%)
Length = 743
Score = 32.7 bits (73), Expect = 0.11
Identities = 31/138 (22%), Positives = 56/138 (40%), Gaps = 9/138 (6%)
Frame = +2
Query: 32 FLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVVGLGGLGHMAVKFGKAFGLR 91
F +P + L +C ++V R P ++ ++G G +G + + +AFG
Sbjct: 11 FKLPDNVSLEEGA--MCEPLSVGVHACRRANIGPETNVLIMGAGPIGLVTMLAARAFGAP 184
Query: 92 VTVFSTSMSKKEEALSFLGADQFV-VSSNQEEM--------RALAKTLDFIIDTASGDHP 142
TV + LGAD + VS+N +++ + + +D D A D
Sbjct: 185 KTVIVDVDDHRLSVAKSLGADDIIKVSTNIKDVAEEVVQIQKVMGAGIDVTFDCAGFDKT 364
Query: 143 FDPYMSLLKISGVLALVG 160
+S + G + LVG
Sbjct: 365 MSTALSATQPGGKVCLVG 418
>BE822669 weakly similar to GP|17978983|gb At1g79870/F19K16_17 {Arabidopsis
thaliana}, partial (56%)
Length = 626
Score = 32.3 bits (72), Expect = 0.14
Identities = 32/112 (28%), Positives = 47/112 (41%), Gaps = 17/112 (15%)
Frame = -2
Query: 66 GKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSF------LGADQ---FVV 116
GK++G+VGLG +G K + FG V+ S S K E + L A+ FV
Sbjct: 574 GKAVGIVGLGRIGWAIAKRAEGFGCPVSYHSRS-EKSETGYKYYSHIIDLAANSEVLFVA 398
Query: 117 SSNQEEMRALAK--TLD------FIIDTASGDHPFDPYMSLLKISGVLALVG 160
+ EE R + +D +I+ G H +P + I G L G
Sbjct: 397 CTLSEETRHIVNRGVIDALGPKGILINVGRGPHVDEPELVAALIEGRLGGAG 242
>TC218190 similar to UP|TIPA_PHAVU (P23958) Probable aquaporin TIP-type alpha
(Tonoplast intrinsic protein alpha) (Alpha TIP), partial
(52%)
Length = 599
Score = 31.2 bits (69), Expect = 0.31
Identities = 19/65 (29%), Positives = 34/65 (52%), Gaps = 2/65 (3%)
Frame = +2
Query: 57 MMRHNMNQPGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLG--ADQF 114
++ NM G S+ + GLG + ++ FGL TV++T++ K ++ + A F
Sbjct: 32 LVTXNMRPQGFSVSI-GLGAFHGLVLEIALTFGLMYTVYATAIDPKRGSIGSIAPLAIGF 208
Query: 115 VVSSN 119
VV +N
Sbjct: 209 VVGAN 223
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,587,355
Number of Sequences: 63676
Number of extensions: 97755
Number of successful extensions: 498
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 495
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 495
length of query: 194
length of database: 12,639,632
effective HSP length: 92
effective length of query: 102
effective length of database: 6,781,440
effective search space: 691706880
effective search space used: 691706880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC123898.9


