

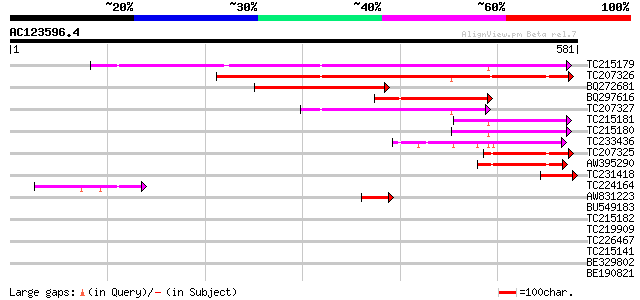
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123596.4 - phase: 0
(581 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC215179 homologue to UP|Q94IR2 (Q94IR2) Carotenoid 9,10-9',10' ... 354 7e-98
TC207326 homologue to UP|Q9FS24 (Q9FS24) Neoxanthin cleavage enz... 293 1e-79
BQ272681 similar to GP|22335707|db nine-cis-epoxycarotenoid diox... 237 1e-62
BQ297616 similar to GP|2828292|emb neoxanthin cleavage enzyme-li... 187 1e-47
TC207327 homologue to UP|Q9M6E8 (Q9M6E8) 9-cis-epoxycarotenoid d... 144 1e-34
TC215181 homologue to UP|Q8LP17 (Q8LP17) Nine-cis-epoxycarotenoi... 97 2e-20
TC215180 homologue to UP|Q8LP17 (Q8LP17) Nine-cis-epoxycarotenoi... 94 2e-19
TC233436 weakly similar to UP|Q8LP17 (Q8LP17) Nine-cis-epoxycaro... 93 3e-19
TC207325 homologue to UP|Q9FS24 (Q9FS24) Neoxanthin cleavage enz... 89 4e-18
AW395290 similar to GP|22335703|dbj nine-cis-epoxycarotenoid dio... 82 5e-16
TC231418 similar to UP|Q8LP14 (Q8LP14) Nine-cis-epoxycarotenoid ... 61 1e-09
TC224164 similar to UP|Q9M6E8 (Q9M6E8) 9-cis-epoxycarotenoid dio... 56 5e-08
AW831223 homologue to GP|22335695|dbj nine-cis-epoxycarotenoid d... 42 8e-04
BU549183 similar to GP|18182311|gb GT-2 factor {Glycine max}, pa... 39 0.005
TC215182 homologue to UP|Q94IR2 (Q94IR2) Carotenoid 9,10-9',10' ... 39 0.008
TC219909 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-... 37 0.019
TC226467 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 37 0.019
TC215141 similar to GB|AAL32703.1|17065098|AY062625 nucleotide s... 37 0.019
BE329802 similar to GP|12081942|dbj nucleotide pyrophosphatase-l... 37 0.019
BE190821 homologue to GP|20160466|dbj B1046G12.17 {Oryza sativa ... 37 0.024
>TC215179 homologue to UP|Q94IR2 (Q94IR2) Carotenoid 9,10-9',10' cleavage
dioxygenase, complete
Length = 1934
Score = 354 bits (908), Expect = 7e-98
Identities = 202/513 (39%), Positives = 291/513 (56%), Gaps = 20/513 (3%)
Frame = +1
Query: 83 DIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQ 142
D++ + + S P H L+ NFAPV E PPT+ +KG LP LNG ++R GPNP+
Sbjct: 199 DLLEKLVVKFLYDSSLPHHYLTGNFAPV-SETPPTKDLPVKGYLPDCLNGEFVRVGPNPK 375
Query: 143 FLPRGPYHLFDGDGMLHAITISNGKATLCSRYVQTYKYKIENEAGYQLIPNV--FSGFNS 200
F P YH FDGDGM+H + I +GKAT SR+V+T + K E G + G
Sbjct: 376 FAPVAGYHWFDGDGMIHGLRIKDGKATYVSRFVRTSRLKQEEYFGGSKFMKIGDLKGLFG 555
Query: 201 LIASAARGSVTAARVISGQYNPSNGIGLANTSLALFGNKLFALGESDLPYEINLTPNGDI 260
L+ T +V+ Y G G ANT+L KL AL E+D PY I + +GD+
Sbjct: 556 LLMVNIHMLRTKWKVLDASY----GTGTANTALVYHHGKLLALSEADKPYAIKVFEDGDL 723
Query: 261 QTIGRYDFNGKLSMSMTAHPKIDGDTGETFAFRYGPMPPFLTYFRFDANGVKHNDVPVFS 320
QT+G D++ +L S TAHPK+D TGE F F Y PP++TY +G H+ VP+ +
Sbjct: 724 QTLGMLDYDKRLGHSFTAHPKVDPFTGEMFTFGYAHTPPYITYRVISKDGYMHDPVPI-T 900
Query: 321 MTRPSFLHDFAITKKYAVFTDIQLGMNPLDMISGGSPVGS-DPSKVSRIGILPRYASDES 379
++ P +HDFAIT+ YA+F D+ L P +M+ + + S D +K +R G+LPRYA DE
Sbjct: 901 VSDPIMMHDFAITENYAIFLDLPLIFRPKEMVKNKTLIFSFDSTKKARFGVLPRYAKDEK 1080
Query: 380 KMKWFDVPGFNIMHAINSWDGEDDEETVT--LIAPNVLSIEHT-MERLELVHAMIEKVKI 436
++WF++P I H N+W+ ED+ +T L PN+ + T E+LE + +++
Sbjct: 1081LIRWFELPNCFIFHNANAWEEEDEVVLITCRLQNPNLDLVGGTAKEKLENFSNELYEMRF 1260
Query: 437 NIKTGIVSRQPLSARNLDFAVINGDYMGKKNRFVYAAIGNPMPKISGVVKIDV------- 489
N+KTG S++ LSA +DF +N Y G+K R+VY + + K++G++K D+
Sbjct: 1261NMKTGEASQKKLSASAVDFPRVNESYTGRKQRYVYGTTLDSIAKVTGIIKFDLHAEPDNG 1440
Query: 490 -----LKGEEVGCRLYGEGCYGGEPFFVAREDG--EEEDDGYLVSYVHDEKKGESKFLVM 542
+ G G G G YG E +V R G EEDDGYL+ +VHDE G+S V+
Sbjct: 1441KTKLEVGGNVQGLYDLGPGKYGSEAVYVPRVPGTDSEEDDGYLICFVHDENTGKSFVHVI 1620
Query: 543 DAKSPEFEIVAEVKLPRRVPYGFHGLFVKESDI 575
+AK+ + VA V+LP RVPYGFH FV E +
Sbjct: 1621NAKTMSADPVAVVELPHRVPYGFHAFFVTEEQL 1719
>TC207326 homologue to UP|Q9FS24 (Q9FS24) Neoxanthin cleavage enzyme, partial
(63%)
Length = 1509
Score = 293 bits (750), Expect = 1e-79
Identities = 151/370 (40%), Positives = 231/370 (61%), Gaps = 5/370 (1%)
Frame = +2
Query: 213 ARVISGQYNPSNGIGLANTSLALFGNKLFALGESDLPYEINLTPNGDIQTIGRYDFNGKL 272
AR + G + S+G+G+AN L F N L A+ E DLPY + +TPNGD+ T+GRY+FNG+L
Sbjct: 65 ARSLFGLVDGSHGMGVANAGLVYFNNHLLAMSEDDLPYHLRITPNGDLTTVGRYNFNGQL 244
Query: 273 SMSMTAHPKIDGDTGETFAFRYGPM-PPFLTYFRFDANGVKHNDVPVFSMTRPSFLHDFA 331
+M AHPK+D T A Y + P+L YFRF +GVK DV + + P+ +HDFA
Sbjct: 245 KSTMIAHPKLDPVTNNLHALSYDVVQKPYLKYFRFSPDGVKSPDVEI-PLKEPTMMHDFA 421
Query: 332 ITKKYAVFTDIQLGMNPLDMISGGSPVGSDPSKVSRIGILPRYASDESKMKWFDVPGFNI 391
IT+ + V D Q+ +MI+GGSPV D +KVSR GIL + A D + MKW D P
Sbjct: 422 ITENFVVVPDQQVVFKLSEMITGGSPVVYDKNKVSRFGILDKNAKDANDMKWIDAPECFC 601
Query: 392 MHAINSWDGEDDEETVTLIAPNVLSIEHTMERLELVHAMIEKVKINIKTGIVSRQPLSAR 451
H N+W+ +++E V + + + E E + +++ ++++N+KTG +R+P+ +
Sbjct: 602 FHLWNAWEEPENDEIVVIGSCMTPADSIFNECEESLKSILSEIRLNLKTGKSTRKPIISE 781
Query: 452 ----NLDFAVINGDYMGKKNRFVYAAIGNPMPKISGVVKIDVLKGEEVGCRLYGEGCYGG 507
NL+ ++N + +G+K +F Y A+ P PK+SG K+D+ G EV +YGE +GG
Sbjct: 782 SEQVNLEAGMVNRNKLGRKTKFGYLALAEPWPKVSGFAKVDLFSG-EVKKYMYGEERFGG 958
Query: 508 EPFFVAREDGEEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLPRRVPYGFHG 567
EP F+ +EDDG+++++VHDEK+ +S+ +++AK+ + E A VKLP RVPYGFHG
Sbjct: 959 EPLFLPNGVDGDEDDGHILAFVHDEKEWKSELQIVNAKTLKLE--ASVKLPSRVPYGFHG 1132
Query: 568 LFVKESDITK 577
F+ D+ K
Sbjct: 1133TFIHSKDLRK 1162
>BQ272681 similar to GP|22335707|db nine-cis-epoxycarotenoid dioxygenase4
{Pisum sativum}, partial (23%)
Length = 421
Score = 237 bits (604), Expect = 1e-62
Identities = 110/138 (79%), Positives = 122/138 (87%)
Frame = +3
Query: 252 INLTPNGDIQTIGRYDFNGKLSMSMTAHPKIDGDTGETFAFRYGPMPPFLTYFRFDANGV 311
+N+TP+GDI+T+GR+DF+GKL+ SMTAHPKID DT E FAFRYGP+PPFLTYFRFD NG
Sbjct: 6 VNVTPDGDIETLGRHDFDGKLTFSMTAHPKIDPDTAECFAFRYGPVPPFLTYFRFDGNGK 185
Query: 312 KHNDVPVFSMTRPSFLHDFAITKKYAVFTDIQLGMNPLDMISGGSPVGSDPSKVSRIGIL 371
KH DVP+FSM PSFLHDFAITKKYA+F DIQLGMNPLDMISGGSPVGS SKV RIGIL
Sbjct: 186 KHEDVPIFSMLTPSFLHDFAITKKYAIFCDIQLGMNPLDMISGGSPVGSVASKVPRIGIL 365
Query: 372 PRYASDESKMKWFDVPGF 389
PR A DES MKWF+VPGF
Sbjct: 366 PRDAKDESMMKWFEVPGF 419
>BQ297616 similar to GP|2828292|emb neoxanthin cleavage enzyme-like protein
{Arabidopsis thaliana}, partial (19%)
Length = 431
Score = 187 bits (475), Expect = 1e-47
Identities = 86/120 (71%), Positives = 108/120 (89%)
Frame = -2
Query: 375 ASDESKMKWFDVPGFNIMHAINSWDGEDDEETVTLIAPNVLSIEHTMERLELVHAMIEKV 434
A DES MKWF+VPGFNI+HAIN+W+ ED+ TV L+APN+LS+EH +ER+ELVHAM+EK+
Sbjct: 370 AKDESMMKWFEVPGFNIIHAINAWE-EDEGRTVVLVAPNILSMEHALERMELVHAMVEKI 194
Query: 435 KINIKTGIVSRQPLSARNLDFAVINGDYMGKKNRFVYAAIGNPMPKISGVVKIDVLKGEE 494
+I + TGI++RQP+SARNLDFAVIN ++GKKNRFVYAA+G+PMPKISGVVK+DV KGEE
Sbjct: 193 RIELDTGIITRQPVSARNLDFAVINPAFVGKKNRFVYAAVGDPMPKISGVVKLDVSKGEE 14
>TC207327 homologue to UP|Q9M6E8 (Q9M6E8) 9-cis-epoxycarotenoid dioxygenase,
partial (32%)
Length = 599
Score = 144 bits (362), Expect = 1e-34
Identities = 74/198 (37%), Positives = 118/198 (59%), Gaps = 4/198 (2%)
Frame = +1
Query: 299 PFLTYFRFDANGVKHNDVPVFSMTRPSFLHDFAITKKYAVFTDIQLGMNPLDMISGGSPV 358
P+L YFRF NGVK DV + + P+ +HDFAIT+ + V D Q+ +MISGGSPV
Sbjct: 7 PYLKYFRFSPNGVKSPDVEI-PLKEPTMMHDFAITENFVVIPDQQVVFKLSEMISGGSPV 183
Query: 359 GSDPSKVSRIGILPRYASDESKMKWFDVPGFNIMHAINSWDGEDDEETVTLIAPNVLSIE 418
D +KVSR GIL + A D + MKW D P H N+W+ +++E V + + +
Sbjct: 184 VYDKNKVSRFGILNKNAKDPNGMKWIDAPECFCFHLWNAWEEPENDEIVVIGSCMTPADS 363
Query: 419 HTMERLELVHAMIEKVKINIKTGIVSRQPLSAR----NLDFAVINGDYMGKKNRFVYAAI 474
E E + +++ ++++N+KTG +R+P+ + NL+ ++N + +G+K +F Y A+
Sbjct: 364 IFNECDESLKSVLSEIRLNLKTGKSTRKPIISESQQVNLEAGMVNRNKLGRKTQFAYLAL 543
Query: 475 GNPMPKISGVVKIDVLKG 492
P PK+SG K+D+ G
Sbjct: 544 AEPWPKVSGFAKVDLFSG 597
>TC215181 homologue to UP|Q8LP17 (Q8LP17) Nine-cis-epoxycarotenoid
dioxygenase1, partial (25%)
Length = 741
Score = 97.1 bits (240), Expect = 2e-20
Identities = 53/135 (39%), Positives = 74/135 (54%), Gaps = 14/135 (10%)
Frame = +2
Query: 455 FAVINGDYMGKKNRFVYAAIGNPMPKISGVVKIDV------------LKGEEVGCRLYGE 502
F +N Y G+K R+VY + + K++G++K D+ + G G G
Sbjct: 146 FGKVNESYTGRKQRYVYGTTLDSIAKVTGIIKFDLHAEPDNGKTKLEVGGNVQGLYDLGP 325
Query: 503 GCYGGEPFFVAREDG--EEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLPRR 560
G YG E +V R G EEDDGYL+ +VHDE G+S V++AK+ + VA V+LP R
Sbjct: 326 GKYGSEAVYVPRVPGTDSEEDDGYLICFVHDENTGKSFVHVINAKTMSADPVAVVELPHR 505
Query: 561 VPYGFHGLFVKESDI 575
VPYGFH FV E +
Sbjct: 506 VPYGFHAFFVTEEQL 550
>TC215180 homologue to UP|Q8LP17 (Q8LP17) Nine-cis-epoxycarotenoid
dioxygenase1, partial (26%)
Length = 708
Score = 94.0 bits (232), Expect = 2e-19
Identities = 52/137 (37%), Positives = 74/137 (53%), Gaps = 14/137 (10%)
Frame = +1
Query: 453 LDFAVINGDYMGKKNRFVYAAIGNPMPKISGVVKIDV------------LKGEEVGCRLY 500
+DF +N Y G+K R+VY + + K++G++K D+ + G G
Sbjct: 1 VDFPRVNESYTGRKQRYVYGTTLDSIAKVTGIIKFDLHAEPDHGKTKLEVGGNVQGLYDL 180
Query: 501 GEGCYGGEPFFVAREDG--EEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLP 558
G G YG E +V R G EEDDGYL+ + HDE +S V++AK+ + VA V+LP
Sbjct: 181 GPGKYGSEAVYVPRVPGTDSEEDDGYLIFFAHDENTRKSFVHVINAKTMSADPVAVVELP 360
Query: 559 RRVPYGFHGLFVKESDI 575
RVPYGFH FV E +
Sbjct: 361 HRVPYGFHAFFVTEEQL 411
>TC233436 weakly similar to UP|Q8LP17 (Q8LP17) Nine-cis-epoxycarotenoid
dioxygenase1, partial (9%)
Length = 806
Score = 93.2 bits (230), Expect = 3e-19
Identities = 61/207 (29%), Positives = 109/207 (52%), Gaps = 29/207 (14%)
Frame = +1
Query: 393 HAINSWDGEDDEETVTLIAPNVLSI----EHTMERLELVHAMIEKVKINIKTGIVSRQPL 448
H INS+ ED E V ++ S+ + +++ E + + + ++N++TG V + L
Sbjct: 22 HIINSF--EDGHEVVVRGCRSLDSLIPGPDPSLKEFEWL-SRCHEWRLNMQTGEVKERGL 192
Query: 449 SARNL---DFAVINGDYMGKKNRFVYAAIGNPM-------PKISGVVKIDV--------L 490
N+ DF +ING+++G +NR+ Y + +P+ PK G+ K+ +
Sbjct: 193 CGANIVYMDFPMINGNFIGIRNRYAYTQVVDPIASSTQDVPKYGGLAKLYFEESCAKFSM 372
Query: 491 KGEE-------VGCRLYGEGCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESKFLVMD 543
+ E V C ++ + + FV R+ G EEDDG+++++VH+E + ++D
Sbjct: 373 RNREQPEEPIRVECHMFEKNTFCSGAAFVPRDGGLEEDDGWIIAFVHNEDTNICEVHIID 552
Query: 544 AKSPEFEIVAEVKLPRRVPYGFHGLFV 570
K E VA++ +PRRVPYGFHG F+
Sbjct: 553 TKKFSGETVAKITMPRRVPYGFHGAFM 633
>TC207325 homologue to UP|Q9FS24 (Q9FS24) Neoxanthin cleavage enzyme, partial
(14%)
Length = 608
Score = 89.4 bits (220), Expect = 4e-18
Identities = 43/92 (46%), Positives = 64/92 (68%)
Frame = +2
Query: 486 KIDVLKGEEVGCRLYGEGCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESKFLVMDAK 545
K+D+ GE V +YGE +GGEP F+ +EDDGY++++VHDEK+ +S+ +++AK
Sbjct: 5 KVDLFSGE-VNKYMYGEERFGGEPLFLPNGVDGDEDDGYILAFVHDEKEWKSELQIVNAK 181
Query: 546 SPEFEIVAEVKLPRRVPYGFHGLFVKESDITK 577
+ + E A VKLP RVPYGFHG F+ +D+ K
Sbjct: 182 TLKLE--ASVKLPSRVPYGFHGTFIHSNDLRK 271
>AW395290 similar to GP|22335703|dbj nine-cis-epoxycarotenoid dioxygenase3
{Pisum sativum}, partial (14%)
Length = 396
Score = 82.4 bits (202), Expect = 5e-16
Identities = 44/93 (47%), Positives = 61/93 (65%), Gaps = 1/93 (1%)
Frame = +1
Query: 480 KISGVVKIDVLKGEEVGCRLYGEGCYGGEPFFVAREDGE-EEDDGYLVSYVHDEKKGESK 538
K+SGV K+D+ GE V YGE +GGEPFF+ G ED+GY++++VHDE +S+
Sbjct: 13 KVSGVAKVDLESGE-VKRHEYGERRFGGEPFFLPTRGGNGNEDEGYVMAFVHDEMTWQSE 189
Query: 539 FLVMDAKSPEFEIVAEVKLPRRVPYGFHGLFVK 571
+++A + E A V LP RVPYGFHG FV+
Sbjct: 190 LQILNALDLKLE--ATVMLPSRVPYGFHGTFVE 282
>TC231418 similar to UP|Q8LP14 (Q8LP14) Nine-cis-epoxycarotenoid
dioxygenase4, partial (6%)
Length = 490
Score = 61.2 bits (147), Expect = 1e-09
Identities = 26/37 (70%), Positives = 34/37 (91%)
Frame = +1
Query: 545 KSPEFEIVAEVKLPRRVPYGFHGLFVKESDITKLSVS 581
+SPE ++VA V+LPRRVPYGFHGLFVKES++ K+S+S
Sbjct: 13 RSPELDVVAAVRLPRRVPYGFHGLFVKESELRKVSLS 123
>TC224164 similar to UP|Q9M6E8 (Q9M6E8) 9-cis-epoxycarotenoid dioxygenase,
partial (22%)
Length = 557
Score = 55.8 bits (133), Expect = 5e-08
Identities = 38/122 (31%), Positives = 54/122 (44%), Gaps = 7/122 (5%)
Frame = +2
Query: 26 SIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTTHNLKKPQTSS---LPALLFNTFD 82
S++T P+ Q + TTTTT T T P I + K P S L + D
Sbjct: 194 SLQTLHFPKKYQPTSTTTTTTTTTTPARETKPVIASPSETKHPLPQSWNFLQKAAASALD 373
Query: 83 DIINTFIDP----PIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNG 138
+ + P+ + DPR ++ NFAPV E P + G +P + GVY+RNG
Sbjct: 374 MVETALVSHERKHPLPKTADPRVQIAGNFAPV-PEHPAQHSLPVAGKIPKCIEGVYVRNG 550
Query: 139 PN 140
N
Sbjct: 551 AN 556
>AW831223 homologue to GP|22335695|dbj nine-cis-epoxycarotenoid dioxygenase1
{Pisum sativum}, partial (8%)
Length = 419
Score = 42.0 bits (97), Expect = 8e-04
Identities = 16/33 (48%), Positives = 23/33 (69%)
Frame = +2
Query: 361 DPSKVSRIGILPRYASDESKMKWFDVPGFNIMH 393
D +K +R G+LPRYA DE ++WF++P I H
Sbjct: 179 DSTKKARFGVLPRYAKDEKLIRWFELPNCFIFH 277
>BU549183 similar to GP|18182311|gb GT-2 factor {Glycine max}, partial (58%)
Length = 512
Score = 39.3 bits (90), Expect = 0.005
Identities = 35/103 (33%), Positives = 44/103 (41%), Gaps = 5/103 (4%)
Frame = +1
Query: 8 ITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPT--KTLPTPPTITTTHNL 65
I KQ PT PP +T T S + TTTTTP + LP PP +N
Sbjct: 163 IHGKQSPTPPKPPP------QTTALSATPVSIVVTTTTTTPSSIIMPLPLPPVSNNNNNT 324
Query: 66 KKPQTSSLP---ALLFNTFDDIINTFIDPPIKPSVDPRHVLSQ 105
P T+SLP ++L T +N I P PS + L Q
Sbjct: 325 TVPSTTSLPMPQSILNITPPSTLNITI-PSFPPSNPTTYFLPQ 450
>TC215182 homologue to UP|Q94IR2 (Q94IR2) Carotenoid 9,10-9',10' cleavage
dioxygenase, partial (6%)
Length = 546
Score = 38.5 bits (88), Expect = 0.008
Identities = 18/36 (50%), Positives = 24/36 (66%)
Frame = +1
Query: 541 VMDAKSPEFEIVAEVKLPRRVPYGFHGLFVKESDIT 576
V++AK+ + VA V+ P RVPYGFH FV E +T
Sbjct: 1 VINAKTMSADPVAVVEXPHRVPYGFHAFFVTEVCVT 108
>TC219909 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-like protein
2 precursor (Phytocyanin-like protein), partial (21%)
Length = 943
Score = 37.4 bits (85), Expect = 0.019
Identities = 46/179 (25%), Positives = 68/179 (37%), Gaps = 14/179 (7%)
Frame = +1
Query: 6 IIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTTHNL 65
+++T + PPT + PPF KT P SQS T +PP LP+P + +L
Sbjct: 196 VVLTPRTPPTPKT-PPFP----KTSPSPSPSQSPKANPPTVSPP---LPSPSPSVHSKSL 351
Query: 66 KKPQTSSLPALLFNTFDDIIN-------TFIDPP-------IKPSVDPRHVLSQNFAPVL 111
P +S +PA+ I+ T PP PS P + P
Sbjct: 352 -SPASSPVPAVGTPAISPAISIPTLAPETGTPPPSLGPSSSSPPSPGPSSSSPPSPGPSS 528
Query: 112 DELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGMLHAITISNGKATL 170
PP T PPSL P+P+ + + +++ +TI G A L
Sbjct: 529 SSPPPA-----GPTSPPSLAPSSNSTAPSPK---NSGFSVAPSSVLMYCVTIVAGAALL 681
>TC226467 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (6%)
Length = 1017
Score = 37.4 bits (85), Expect = 0.019
Identities = 23/72 (31%), Positives = 30/72 (40%), Gaps = 1/72 (1%)
Frame = +3
Query: 5 PIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLP-TPPTITTTH 63
P I+ K P T P +S PQT Q Q K + PT P PP ++T
Sbjct: 198 PAIVAPKPAPVTPPAPKVAPASSPKAPPPQTPQPQPPKVSPVVTPTSPPPIPPPPVSTPP 377
Query: 64 NLKKPQTSSLPA 75
L P+ + PA
Sbjct: 378 PLPPPKIAPTPA 413
>TC215141 similar to GB|AAL32703.1|17065098|AY062625 nucleotide sugar
epimerase-like protein {Arabidopsis thaliana;} , partial
(89%)
Length = 1743
Score = 37.4 bits (85), Expect = 0.019
Identities = 25/59 (42%), Positives = 30/59 (50%)
Frame = +2
Query: 16 TSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTTHNLKKPQTSSLP 74
+SS PP N SS +S S + TT +PP PTPPT T+T P TSS P
Sbjct: 92 SSSTPPPNSSSAPLSWSHSSSSSSSPSTTLPSPP----PTPPTATST---PTPTTSSPP 247
>BE329802 similar to GP|12081942|dbj nucleotide pyrophosphatase-like protein
{Spinacia oleracea}, partial (17%)
Length = 447
Score = 37.4 bits (85), Expect = 0.019
Identities = 28/75 (37%), Positives = 36/75 (47%), Gaps = 12/75 (16%)
Frame = +1
Query: 10 SKQPPT-----TSSIPPFNISSI--KTKEKPQTSQSQTLK-----TTTTTPPTKTLPTPP 57
S PPT TSS PP SS+ KT +P+ S + TTT + P TLPT
Sbjct: 217 SSFPPTASDSGTSSRPPLPTSSV**KTAPRPRPDSSLSFPPSPSPTTTPSSPGSTLPTTA 396
Query: 58 TITTTHNLKKPQTSS 72
+ TT+ + P SS
Sbjct: 397 SSTTSSSTPSPARSS 441
>BE190821 homologue to GP|20160466|dbj B1046G12.17 {Oryza sativa (japonica
cultivar-group)}, partial (6%)
Length = 473
Score = 37.0 bits (84), Expect = 0.024
Identities = 32/93 (34%), Positives = 41/93 (43%), Gaps = 16/93 (17%)
Frame = +2
Query: 4 KPIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTT----------TPPTKTL 53
KP I + PTTS+ P + S T+ P T++ +TTT TP + T
Sbjct: 182 KPHPIITSFSPTTSNTPTWPPSFATTRSPPTTTRPPLSCSTTTSSSPPPPLPPTPNSSTC 361
Query: 54 P-TPPTITTT-----HNLKKPQTSSLPALLFNT 80
P TPP TT+ NL P S LP L T
Sbjct: 362 PLTPPRQTTSLTPPPSNLSLPTVSLLPFKLSTT 460
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.137 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,978,186
Number of Sequences: 63676
Number of extensions: 401343
Number of successful extensions: 4265
Number of sequences better than 10.0: 283
Number of HSP's better than 10.0 without gapping: 3475
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4001
length of query: 581
length of database: 12,639,632
effective HSP length: 102
effective length of query: 479
effective length of database: 6,144,680
effective search space: 2943301720
effective search space used: 2943301720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC123596.4


