

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
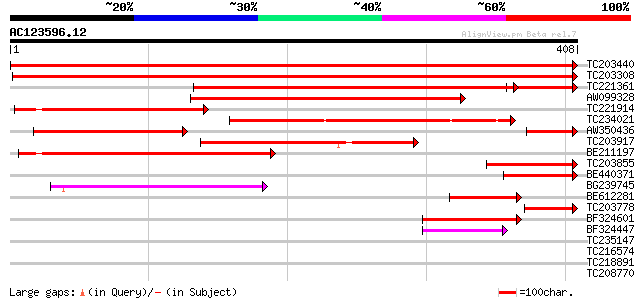
Query= AC123596.12 - phase: 0
(408 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC203440 similar to UP|RL4_PRUAR (Q9XF97) 60S ribosomal protein ... 730 0.0
TC203308 725 0.0
TC221361 similar to UP|RL4_PRUAR (Q9XF97) 60S ribosomal protein ... 387 e-125
AW099328 263 1e-70
TC221914 similar to UP|RL4_PRUAR (Q9XF97) 60S ribosomal protein ... 238 5e-63
TC234021 weakly similar to UP|Q8LA93 (Q8LA93) 60S ribosomal prot... 228 4e-60
AW350436 homologue to SP|Q9XF97|RL4_ 60S ribosomal protein L4 (L... 211 6e-55
TC203917 similar to UP|RL4_PRUAR (Q9XF97) 60S ribosomal protein ... 204 4e-53
BE211197 weakly similar to SP|Q9XF97|RL4_ 60S ribosomal protein ... 177 8e-45
TC203855 weakly similar to UP|Q6VVA6 (Q6VVA6) KUP1, partial (11%) 119 2e-27
BE440371 similar to SP|Q9XF97|RL4_P 60S ribosomal protein L4 (L1... 97 2e-20
BG239745 similar to SP|Q9XF97|RL4_P 60S ribosomal protein L4 (L1... 94 1e-19
BE612281 similar to GP|21593543|gb| 60S ribosomal protein L4-B (... 81 1e-15
TC203778 similar to UP|RL4_PRUAR (Q9XF97) 60S ribosomal protein ... 75 4e-14
BF324601 similar to GP|17862306|gb| LD21756p {Drosophila melanog... 65 4e-11
BF324447 weakly similar to GP|17862306|gb| LD21756p {Drosophila ... 54 1e-07
TC235147 weakly similar to UP|Q6NAC8 (Q6NAC8) Possible cobalamin... 39 0.004
TC216574 similar to UP|RK4_SPIOL (O49937) 50S ribosomal protein ... 35 0.061
TC218891 homologue to UP|GAC1_YEAST (P28006) Protein phosphatase... 32 0.67
TC208770 weakly similar to UP|Q84ME1 (Q84ME1) At5g19080/T16G12_1... 31 1.1
>TC203440 similar to UP|RL4_PRUAR (Q9XF97) 60S ribosomal protein L4 (L1),
complete
Length = 1576
Score = 730 bits (1885), Expect = 0.0
Identities = 362/408 (88%), Positives = 388/408 (94%)
Frame = +1
Query: 1 MAAAAAAARPLVTIQTLESEMSTDSPNTLPIPDVMRASIRPDIVNFVHSNISKNSRQPYA 60
+ A AAAARPLVT+Q L+ +M+TD+ T+PIPDVM+ASIRPDIVNFVHSNISKNSRQPYA
Sbjct: 148 LTAMAAAARPLVTVQALDGDMATDASATVPIPDVMKASIRPDIVNFVHSNISKNSRQPYA 327
Query: 61 VSRKAGHQTSAESWGTGRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHR 120
VSR+AGHQTSAESWGTGRAVSRIPRV GGGTHRAGQ AFGNMCRGGRMFAPT+IWR+WHR
Sbjct: 328 VSRRAGHQTSAESWGTGRAVSRIPRVPGGGTHRAGQGAFGNMCRGGRMFAPTKIWRRWHR 507
Query: 121 KINVNQKRYAVVSAIAASAIPSLVLARGHRIETVPEFPLVVGDAAEGVEKTKEAIKVLKQ 180
KINVNQKRYAVVSAIAASAIPSLVLARGHRIE+VPE PLVV D+AEGVEKTKEAIKVLKQ
Sbjct: 508 KINVNQKRYAVVSAIAASAIPSLVLARGHRIESVPELPLVVSDSAEGVEKTKEAIKVLKQ 687
Query: 181 VGAFADAEKAKDSLGIRPGKGKMRNRRYISRKGPLIVYGTDGAKAVKAFRNIPGVEITNV 240
+GAF DAEKAKDS IRPGKGKMRNRRYISR+GPL+VYGT+GAKAVKAFRN+PGVE+ NV
Sbjct: 688 IGAFPDAEKAKDSRSIRPGKGKMRNRRYISRRGPLVVYGTEGAKAVKAFRNVPGVEVANV 867
Query: 241 ERLNLLKLAPGGHLGRFVIWTKSAFEKLDSIYGTFDKGSEKKKGYVLPRAKMVNSDLTRI 300
ERLNLLKLAPGGHLGRFVIWTKSAFEKLDSIYG+FDK SEKKKGY+LPR+KMVNSDL+RI
Sbjct: 868 ERLNLLKLAPGGHLGRFVIWTKSAFEKLDSIYGSFDKPSEKKKGYLLPRSKMVNSDLSRI 1047
Query: 301 INSDEVQSVVRPIKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKSKKE 360
INSDEVQSVVRPIKKDVKRATLKKNPLKNLN ML+LNPYAKTAKRMALLAEA+RVK KKE
Sbjct: 1048INSDEVQSVVRPIKKDVKRATLKKNPLKNLNSMLKLNPYAKTAKRMALLAEAQRVKVKKE 1227
Query: 361 KFDKKRKIVSKEEASAIKAAGKAWYNTMVSDSDYAEFDNFSKWLGVSQ 408
K DKKRK VSKEEASAIKAAGKAWY+TMVSDSDY EFDNFSKWLGVSQ
Sbjct: 1228KLDKKRKTVSKEEASAIKAAGKAWYHTMVSDSDYTEFDNFSKWLGVSQ 1371
>TC203308
Length = 1778
Score = 725 bits (1872), Expect = 0.0
Identities = 359/406 (88%), Positives = 386/406 (94%)
Frame = -3
Query: 3 AAAAAARPLVTIQTLESEMSTDSPNTLPIPDVMRASIRPDIVNFVHSNISKNSRQPYAVS 62
A AAAARPLVT+Q L+S+M+TD+ T+PIPDVM++SIRPDIVNFVHSNISKNSRQPYAVS
Sbjct: 1728 AMAAAARPLVTVQALDSDMATDASATVPIPDVMKSSIRPDIVNFVHSNISKNSRQPYAVS 1549
Query: 63 RKAGHQTSAESWGTGRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHRKI 122
R+AGHQTSAESWGTGRAVSRIPRV GGGTHRAGQ AFGNMCRGGRMFAPT+IWR+WHRKI
Sbjct: 1548 RRAGHQTSAESWGTGRAVSRIPRVPGGGTHRAGQGAFGNMCRGGRMFAPTKIWRRWHRKI 1369
Query: 123 NVNQKRYAVVSAIAASAIPSLVLARGHRIETVPEFPLVVGDAAEGVEKTKEAIKVLKQVG 182
NVNQKRYAVVSAIAASAIPSLVLARGHRIE+VPE PLVV D+AE VEKTKEAIKVLKQ+G
Sbjct: 1368 NVNQKRYAVVSAIAASAIPSLVLARGHRIESVPELPLVVSDSAESVEKTKEAIKVLKQIG 1189
Query: 183 AFADAEKAKDSLGIRPGKGKMRNRRYISRKGPLIVYGTDGAKAVKAFRNIPGVEITNVER 242
AF DAEKAKDS IRPGKGKMRNRRYISR+GPLIVYGT+GAKAVKAFRN+PGVE+ NVER
Sbjct: 1188 AFPDAEKAKDSRSIRPGKGKMRNRRYISRRGPLIVYGTEGAKAVKAFRNVPGVEVANVER 1009
Query: 243 LNLLKLAPGGHLGRFVIWTKSAFEKLDSIYGTFDKGSEKKKGYVLPRAKMVNSDLTRIIN 302
LNLLKLAPGGHLGRFV+WTKSAFEKLDSIYG+FDK SEKKKGY+LPR KMVNSDL RIIN
Sbjct: 1008 LNLLKLAPGGHLGRFVVWTKSAFEKLDSIYGSFDKPSEKKKGYLLPRPKMVNSDLARIIN 829
Query: 303 SDEVQSVVRPIKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKSKKEKF 362
SDEVQSVVRPIKKDVK+ATLKKNPLKNLN ML+LNPYAKTAKRMALLAEA+RVK+KKEK
Sbjct: 828 SDEVQSVVRPIKKDVKKATLKKNPLKNLNSMLKLNPYAKTAKRMALLAEAQRVKAKKEKL 649
Query: 363 DKKRKIVSKEEASAIKAAGKAWYNTMVSDSDYAEFDNFSKWLGVSQ 408
DKKRK VSKEEASAIK+AGKAWY+TMVSDSDY EFDNFSKWLGVSQ
Sbjct: 648 DKKRKTVSKEEASAIKSAGKAWYHTMVSDSDYTEFDNFSKWLGVSQ 511
>TC221361 similar to UP|RL4_PRUAR (Q9XF97) 60S ribosomal protein L4 (L1),
partial (67%)
Length = 878
Score = 387 bits (994), Expect(2) = e-125
Identities = 194/234 (82%), Positives = 213/234 (90%)
Frame = +1
Query: 133 SAIAASAIPSLVLARGHRIETVPEFPLVVGDAAEGVEKTKEAIKVLKQVGAFADAEKAKD 192
SAIAASAIPSLV ARGHRIE+VPE PLVV D E VEK+KEA+K+L+++GAF DAEKAK
Sbjct: 1 SAIAASAIPSLVQARGHRIESVPELPLVVSDTVESVEKSKEAVKILQKIGAFPDAEKAKL 180
Query: 193 SLGIRPGKGKMRNRRYISRKGPLIVYGTDGAKAVKAFRNIPGVEITNVERLNLLKLAPGG 252
S GIRPGKGKMRNRR+ISRKGPLIVYGT+GAK VKAFRNIPGVE+ NV+RLNLLKLAPGG
Sbjct: 181 SHGIRPGKGKMRNRRHISRKGPLIVYGTEGAKVVKAFRNIPGVEVANVDRLNLLKLAPGG 360
Query: 253 HLGRFVIWTKSAFEKLDSIYGTFDKGSEKKKGYVLPRAKMVNSDLTRIINSDEVQSVVRP 312
HLGRF+IWTKSAFEKLDSIYG+F K SEKKKGY+LPR +MVNSDL RIINSDEVQSVVRP
Sbjct: 361 HLGRFIIWTKSAFEKLDSIYGSFHKASEKKKGYLLPRPRMVNSDLARIINSDEVQSVVRP 540
Query: 313 IKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKSKKEKFDKKR 366
IKKDVKRA LKKNPL NLN ML+LNPYAKTAKRMALLAE +R+ +KKEK DKKR
Sbjct: 541 IKKDVKRAPLKKNPLTNLNAMLKLNPYAKTAKRMALLAEKQRIVAKKEKLDKKR 702
Score = 80.9 bits (198), Expect(2) = e-125
Identities = 37/51 (72%), Positives = 43/51 (83%)
Frame = +2
Query: 358 KKEKFDKKRKIVSKEEASAIKAAGKAWYNTMVSDSDYAEFDNFSKWLGVSQ 408
++ + IVSKEEASAIK+AGK+WY TMVSDSDYA+FDNFSKWLGVSQ
Sbjct: 677 RRRSLIRSXXIVSKEEASAIKSAGKSWYQTMVSDSDYADFDNFSKWLGVSQ 829
>AW099328
Length = 631
Score = 263 bits (671), Expect = 1e-70
Identities = 136/198 (68%), Positives = 155/198 (77%)
Frame = +2
Query: 131 VVSAIAASAIPSLVLARGHRIETVPEFPLVVGDAAEGVEKTKEAIKVLKQVGAFADAEKA 190
VVSAIAASAIPSLVL RGHRIE+VPE PLV D+AE VEKTKEA+KVLKQ+GAF DAEKA
Sbjct: 2 VVSAIAASAIPSLVLVRGHRIESVPELPLVASDSAESVEKTKEALKVLKQIGAFPDAEKA 181
Query: 191 KDSLGIRPGKGKMRNRRYISRKGPLIVYGTDGAKAVKAFRNIPGVEITNVERLNLLKLAP 250
KDS IRPGKGKM R YI R+GP+I ++A AFRN+PGVE+ N ERLNLLKLAP
Sbjct: 182 KDSRSIRPGKGKMLYRCYILRRGPIICVRNGRSQADYAFRNVPGVEVANAERLNLLKLAP 361
Query: 251 GGHLGRFVIWTKSAFEKLDSIYGTFDKGSEKKKGYVLPRAKMVNSDLTRIINSDEVQSVV 310
GGHLGRFV+WTKSAFEKLDSI+G+FD SE K Y+LPR KMV DL I++DE QS
Sbjct: 362 GGHLGRFVVWTKSAFEKLDSIHGSFDMPSEMKTAYLLPRPKMVTYDLAMTIHTDEDQSDA 541
Query: 311 RPIKKDVKRATLKKNPLK 328
R I++DV TLK L+
Sbjct: 542 RTIEQDVNNVTLKSTLLR 595
>TC221914 similar to UP|RL4_PRUAR (Q9XF97) 60S ribosomal protein L4 (L1),
partial (33%)
Length = 448
Score = 238 bits (606), Expect = 5e-63
Identities = 115/140 (82%), Positives = 128/140 (91%)
Frame = +2
Query: 4 AAAAARPLVTIQTLESEMSTDSPNTLPIPDVMRASIRPDIVNFVHSNISKNSRQPYAVSR 63
A AAARP+V++QTLE D+ T+P+PDVM+ASIRPDIVNFVHSNIS+NSRQPYAVS+
Sbjct: 41 AVAAARPIVSVQTLEG----DATPTVPLPDVMKASIRPDIVNFVHSNISRNSRQPYAVSK 208
Query: 64 KAGHQTSAESWGTGRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHRKIN 123
+AGHQTSAESWGTGRAVSRIPRV GGGTHRAGQ FGNMCRGGRMFAPT+IWR+WHRKIN
Sbjct: 209 RAGHQTSAESWGTGRAVSRIPRVPGGGTHRAGQGTFGNMCRGGRMFAPTKIWRRWHRKIN 388
Query: 124 VNQKRYAVVSAIAASAIPSL 143
V QKR+AVVSAIAASAIPSL
Sbjct: 389 VQQKRHAVVSAIAASAIPSL 448
>TC234021 weakly similar to UP|Q8LA93 (Q8LA93) 60S ribosomal protein L4-B
(L1), partial (37%)
Length = 686
Score = 228 bits (581), Expect = 4e-60
Identities = 120/207 (57%), Positives = 153/207 (72%), Gaps = 1/207 (0%)
Frame = +1
Query: 159 LVVGDAA-EGVEKTKEAIKVLKQVGAFADAEKAKDSLGIRPGKGKMRNRRYISRKGPLIV 217
LVV D+A G+EKTK A+ +LK + AF D E +K+S IRPG GK RNRRY+ R+GPL+V
Sbjct: 58 LVVADSAFSGLEKTKHAVALLKALNAFEDVEHSKNSKKIRPGNGKSRNRRYVLRRGPLVV 237
Query: 218 YGTDGAKAVKAFRNIPGVEITNVERLNLLKLAPGGHLGRFVIWTKSAFEKLDSIYGTFDK 277
Y G + AFRN+PGVE+ +V RLNLL LAPGGHLGRF+IWTK AF +LDS++GT+ K
Sbjct: 238 YDEAGPLTL-AFRNLPGVEVQHVSRLNLLSLAPGGHLGRFIIWTKGAFARLDSLFGTYKK 414
Query: 278 GSEKKKGYVLPRAKMVNSDLTRIINSDEVQSVVRPIKKDVKRATLKKNPLKNLNVMLRLN 337
+ +K GY LPR K+ N+DL RIINSDEVQ+VVRP++ +T KKNPLKNL +++LN
Sbjct: 415 AAPEKVGYRLPRNKVTNADLARIINSDEVQAVVRPVRPRT-FSTRKKNPLKNLGFLVKLN 591
Query: 338 PYAKTAKRMALLAEAERVKSKKEKFDK 364
PYA KR ALL A++ KKE+ K
Sbjct: 592 PYALAQKRRALLT-AQKKALKKERDPK 669
>AW350436 homologue to SP|Q9XF97|RL4_ 60S ribosomal protein L4 (L1).
[Apricot] {Prunus armeniaca}, partial (35%)
Length = 555
Score = 211 bits (536), Expect = 6e-55
Identities = 97/111 (87%), Positives = 106/111 (95%)
Frame = -3
Query: 18 ESEMSTDSPNTLPIPDVMRASIRPDIVNFVHSNISKNSRQPYAVSRKAGHQTSAESWGTG 77
+ +M+TD+ T+PIPDVM+ASIRPDIVNFVHSNISKNSRQPYAVSR+AGHQTSAESWGTG
Sbjct: 553 DGDMATDASATVPIPDVMKASIRPDIVNFVHSNISKNSRQPYAVSRRAGHQTSAESWGTG 374
Query: 78 RAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHRKINVNQKR 128
RAVSRIPRV GGGTHRAGQ AFGNMCRGGRMFAPT+IWR+WHRKINVNQKR
Sbjct: 373 RAVSRIPRVPGGGTHRAGQGAFGNMCRGGRMFAPTKIWRRWHRKINVNQKR 221
Score = 74.3 bits (181), Expect = 9e-14
Identities = 34/36 (94%), Positives = 35/36 (96%)
Frame = -2
Query: 373 EASAIKAAGKAWYNTMVSDSDYAEFDNFSKWLGVSQ 408
EASAIKAAGKAWY+TMVSDSDY EFDNFSKWLGVSQ
Sbjct: 227 EASAIKAAGKAWYHTMVSDSDYTEFDNFSKWLGVSQ 120
>TC203917 similar to UP|RL4_PRUAR (Q9XF97) 60S ribosomal protein L4 (L1),
partial (35%)
Length = 477
Score = 204 bits (520), Expect = 4e-53
Identities = 109/162 (67%), Positives = 124/162 (76%), Gaps = 5/162 (3%)
Frame = +2
Query: 138 SAIPSLVLARGHRIETVPEFPLVVGDAAEGVEKTKEAIKVLKQVGAFADAEKAKDSLGIR 197
SAIPSLV ARGHRIE+VPE PLVV D E VEK+KEA+K+L+++GAF DAEKAK S GIR
Sbjct: 2 SAIPSLVQARGHRIESVPELPLVVSDTVESVEKSKEAVKILQKIGAFTDAEKAKLSHGIR 181
Query: 198 PGKGKMRNRRYISRKGPLIVYGTDGAKAVKAFRNIPGV-----EITNVERLNLLKLAPGG 252
PGKGKMRNRRYIS KGPLIVY + + V I ++ R KLAPGG
Sbjct: 182 PGKGKMRNRRYISGKGPLIVYXNRRC*GH*SLQEHSXV*RWQTLIGSIRR----KLAPGG 349
Query: 253 HLGRFVIWTKSAFEKLDSIYGTFDKGSEKKKGYVLPRAKMVN 294
HLGRF+IWTKSAFEKLDSIYG+FDK SEKKKGY+LPR KM+N
Sbjct: 350 HLGRFIIWTKSAFEKLDSIYGSFDKASEKKKGYLLPRPKMIN 475
>BE211197 weakly similar to SP|Q9XF97|RL4_ 60S ribosomal protein L4 (L1).
[Apricot] {Prunus armeniaca}, partial (35%)
Length = 544
Score = 177 bits (449), Expect = 8e-45
Identities = 95/185 (51%), Positives = 122/185 (65%)
Frame = +2
Query: 7 AARPLVTIQTLESEMSTDSPNTLPIPDVMRASIRPDIVNFVHSNISKNSRQPYAVSRKAG 66
AARP+V++QTL+ D+ T+P+P+ M+ASIRPDIVNFVHSNIS NS QPYA+ +
Sbjct: 2 AARPIVSVQTLDG----DATPTVPLPNFMKASIRPDIVNFVHSNISHNSHQPYAIIKHTS 169
Query: 67 HQTSAESWGTGRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHRKINVNQ 126
HQTS +S GT +S IP+ THRA Q+AF NM R+FAPT+I H INV Q
Sbjct: 170 HQTSRDS*GTYHTISHIPQFPSKYTHRASQSAFENMYHNNRIFAPTKI*HH*HHNINVQQ 349
Query: 127 KRYAVVSAIAASAIPSLVLARGHRIETVPEFPLVVGDAAEGVEKTKEAIKVLKQVGAFAD 186
K + + SA+AA +PS + AR H I+ V + PLVV DA E VE K+ +KV + G D
Sbjct: 350 KHHTIGSAMAACTMPSRMRARNHHIKCVRKLPLVVSDAVESVENAKKRVKVSMRSGD*HD 529
Query: 187 AEKAK 191
AE AK
Sbjct: 530 AEIAK 544
>TC203855 weakly similar to UP|Q6VVA6 (Q6VVA6) KUP1, partial (11%)
Length = 653
Score = 119 bits (298), Expect = 2e-27
Identities = 58/65 (89%), Positives = 62/65 (95%)
Frame = +3
Query: 344 KRMALLAEAERVKSKKEKFDKKRKIVSKEEASAIKAAGKAWYNTMVSDSDYAEFDNFSKW 403
KRMALLAEA+RVK+KKEK DKKRK VSKEEASAIK+AGKAWY+TMVSDSDY EFDNFSKW
Sbjct: 6 KRMALLAEAQRVKAKKEKLDKKRKTVSKEEASAIKSAGKAWYHTMVSDSDYTEFDNFSKW 185
Query: 404 LGVSQ 408
LGVSQ
Sbjct: 186 LGVSQ 200
>BE440371 similar to SP|Q9XF97|RL4_P 60S ribosomal protein L4 (L1). [Apricot]
{Prunus armeniaca}, partial (12%)
Length = 454
Score = 96.7 bits (239), Expect = 2e-20
Identities = 48/54 (88%), Positives = 50/54 (91%), Gaps = 1/54 (1%)
Frame = +3
Query: 356 KSKKEKF-DKKRKIVSKEEASAIKAAGKAWYNTMVSDSDYAEFDNFSKWLGVSQ 408
K KK+KF DKKRK VSKEEASAIKAAGKAWY+TMVSDSDY EFDNFSKWLGVSQ
Sbjct: 225 KKKKKKFLDKKRKTVSKEEASAIKAAGKAWYHTMVSDSDYTEFDNFSKWLGVSQ 386
>BG239745 similar to SP|Q9XF97|RL4_P 60S ribosomal protein L4 (L1). [Apricot]
{Prunus armeniaca}, partial (17%)
Length = 502
Score = 94.0 bits (232), Expect = 1e-19
Identities = 66/167 (39%), Positives = 87/167 (51%), Gaps = 11/167 (6%)
Frame = +2
Query: 30 PIPDVMRA--------SIRPDIVNFVHSNISKNSRQPYAVSRKAGHQTSAESWGTGRAVS 81
PIPDVM+ +R F + + + +P ++ K + +
Sbjct: 2 PIPDVMKVLHLARTIVKLRTTPTIFKNRPTTPMAVRPPVLATKTSARVLGKPVVPSPVFP 181
Query: 82 RIPRVAGGGTHRAGQAAFGNMCRG-GRMFAPT-RIWRKWHRKINV-NQKRYAVVSAIAAS 138
R+PR A RA MCRG +FAP R +WH + + + VVS IA
Sbjct: 182 RVPRGAEPHPRRAQGVLSETMCRGRNALFAPDQRSGGRWHXENQL*TRSATPVVSGIAGF 361
Query: 139 AIPSLVLARGHRIETVPEFPLVVGDAAEGVEKTKEAIKVLKQVGAFA 185
IP LVLAR HRIE+ PE PLVV D+AE VEKTK+AIKVLKQ+GAF+
Sbjct: 362 VIPFLVLARCHRIESRPELPLVVSDSAESVEKTKKAIKVLKQIGAFS 502
>BE612281 similar to GP|21593543|gb| 60S ribosomal protein L4-B (L1)
{Arabidopsis thaliana}, partial (15%)
Length = 386
Score = 80.9 bits (198), Expect = 1e-15
Identities = 40/52 (76%), Positives = 44/52 (83%)
Frame = +2
Query: 317 VKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKSKKEKFDKKRKI 368
VKRA LKKNPL NLN ML+LNPYAKTAKRMALLAE +R+ +KKEK KKR I
Sbjct: 17 VKRAPLKKNPLTNLNAMLKLNPYAKTAKRMALLAEKQRIVAKKEKLHKKRNI 172
Score = 33.1 bits (74), Expect = 0.23
Identities = 21/73 (28%), Positives = 37/73 (49%)
Frame = +3
Query: 311 RPIKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKSKKEKFDKKRKIVS 370
R +++ + R TL + ++ + + L P + + + ++ F + VS
Sbjct: 12 RGLREHLSRRTLSRTSMRC*SSIPMLRPL----REWLFSLRSSALWLRRRSFIRSATFVS 179
Query: 371 KEEASAIKAAGKA 383
KEEASAIKAAGK+
Sbjct: 180 KEEASAIKAAGKS 218
>TC203778 similar to UP|RL4_PRUAR (Q9XF97) 60S ribosomal protein L4 (L1),
partial (9%)
Length = 919
Score = 75.5 bits (184), Expect = 4e-14
Identities = 34/38 (89%), Positives = 37/38 (96%)
Frame = +1
Query: 371 KEEASAIKAAGKAWYNTMVSDSDYAEFDNFSKWLGVSQ 408
+EEASAIK+AGKAWY+TMVSDSDY EFDNFSKWLGVSQ
Sbjct: 784 QEEASAIKSAGKAWYHTMVSDSDYTEFDNFSKWLGVSQ 897
>BF324601 similar to GP|17862306|gb| LD21756p {Drosophila melanogaster},
partial (6%)
Length = 214
Score = 65.5 bits (158), Expect = 4e-11
Identities = 31/71 (43%), Positives = 46/71 (64%)
Frame = +2
Query: 298 TRIINSDEVQSVVRPIKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKS 357
TR+I S+E++ V+RP +K V R + NP++N MLRLNPYA +R A+L +R+K
Sbjct: 2 TRLIKSEEIREVLRPAQKKVVRRVRRLNPMRNQKAMLRLNPYAAVLRRNAILTIQKRMKQ 181
Query: 358 KKEKFDKKRKI 368
+ E KKR +
Sbjct: 182 RAELLAKKRGV 214
>BF324447 weakly similar to GP|17862306|gb| LD21756p {Drosophila
melanogaster}, partial (15%)
Length = 199
Score = 53.9 bits (128), Expect = 1e-07
Identities = 28/61 (45%), Positives = 37/61 (59%)
Frame = +2
Query: 298 TRIINSDEVQSVVRPIKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKS 357
TR+I SDE+ V+ P K V R + NPL N + MLRLNPYA A+L +R+KS
Sbjct: 2 TRLIKSDEIIKVLMPAHKKVVRRVRRLNPLLNQHAMLRLNPYAAVLTTNAILTIQKRMKS 181
Query: 358 K 358
+
Sbjct: 182 R 184
>TC235147 weakly similar to UP|Q6NAC8 (Q6NAC8) Possible cobalamin
(5`-phosphate) synthase, partial (9%)
Length = 388
Score = 38.9 bits (89), Expect = 0.004
Identities = 21/59 (35%), Positives = 33/59 (55%), Gaps = 3/59 (5%)
Frame = +1
Query: 327 LKNLNVMLRLNPYAKTAKRMALLAEAERVKSKKEKFDKKRKIVS---KEEASAIKAAGK 382
+KNL ++RLNPYA + +R LL + + ++ K + KRK + + A KAA K
Sbjct: 7 VKNLGALVRLNPYALSVRRTELLTQQRQASARAAKIEAKRKNLPAPLTADEKATKAAKK 183
>TC216574 similar to UP|RK4_SPIOL (O49937) 50S ribosomal protein L4,
chloroplast precursor (R-protein L4), partial (74%)
Length = 1364
Score = 35.0 bits (79), Expect = 0.061
Identities = 39/120 (32%), Positives = 53/120 (43%), Gaps = 12/120 (10%)
Frame = +3
Query: 75 GTGRAVSRIPRVAGGGTHRAGQAAFGNMCRG----------GRMFAPTRIWRKWHRKINV 124
GT ++R V GGG Q G RG G +F P R W KIN
Sbjct: 369 GTASTLTR-GEVRGGGRKPFPQKKTGRARRGSNRTPLRPGGGVIFGPKP--RDWSIKINR 539
Query: 125 NQKRYAVVSAIAASAIPSLVLARGHRIETVPEFPLVVGDAAEGVE--KTKEAIKVLKQVG 182
+KR A+ +A+A++A ++V V +F AAE E KTKE I +K+ G
Sbjct: 540 KEKRLAISTAVASAAASAVV---------VEDF------AAEFAEKPKTKEFIAAMKRWG 674
>TC218891 homologue to UP|GAC1_YEAST (P28006) Protein phosphatase 1
regulatory subunit GAC1, partial (3%)
Length = 1463
Score = 31.6 bits (70), Expect = 0.67
Identities = 33/121 (27%), Positives = 47/121 (38%)
Frame = +2
Query: 116 RKWHRKINVNQKRYAVVSAIAASAIPSLVLARGHRIETVPEFPLVVGDAAEGVEKTKEAI 175
+K R+ ++R V++ A IPS+V A G R + G + A
Sbjct: 134 KKDRRRGGYGRRRRDPVASFAGERIPSVV-AEGERSDN-------------GDGRGSGAA 271
Query: 176 KVLKQVGAFADAEKAKDSLGIRPGKGKMRNRRYISRKGPLIVYGTDGAKAVKAFRNIPGV 235
+Q G A A G+GK R RR RKG L+ DG + R + G
Sbjct: 272 TTAEQSGISASDSAA--------GEGKRRRRRRRRRKGGLLERRRDGGADRRVGREVSGT 427
Query: 236 E 236
E
Sbjct: 428 E 430
>TC208770 weakly similar to UP|Q84ME1 (Q84ME1) At5g19080/T16G12_120, partial
(30%)
Length = 753
Score = 30.8 bits (68), Expect = 1.1
Identities = 26/81 (32%), Positives = 39/81 (48%), Gaps = 3/81 (3%)
Frame = -3
Query: 62 SRKAGHQTSAESWGT---GRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKW 118
S ++G + AE WG GRA R AGGG++ AG + GR + R W W
Sbjct: 352 SWRSGGRRRAEEWGQQDEGRATRRAGAGAGGGSNAAGSES------EGREWECRRNW-GW 194
Query: 119 HRKINVNQKRYAVVSAIAASA 139
+ N+K+ + +AA+A
Sbjct: 193 ----SCNRKKLSGFVVVAAAA 143
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,018,035
Number of Sequences: 63676
Number of extensions: 160748
Number of successful extensions: 947
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 939
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 942
length of query: 408
length of database: 12,639,632
effective HSP length: 100
effective length of query: 308
effective length of database: 6,272,032
effective search space: 1931785856
effective search space used: 1931785856
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC123596.12


