

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123593.9 + phase: 0 /pseudo
(409 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
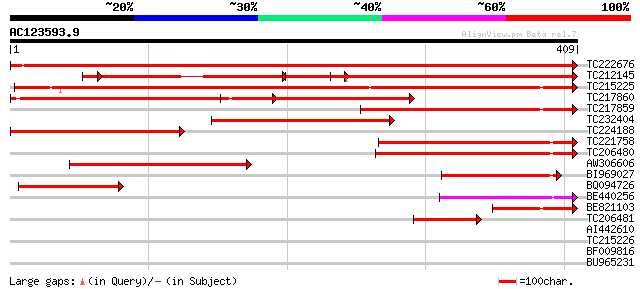
Score E
Sequences producing significant alignments: (bits) Value
TC222676 UP|Q84L88 (Q84L88) Apyrase, complete 710 0.0
TC212145 UP|Q76IB9 (Q76IB9) Apyrase (Fragment), complete 301 e-139
TC215225 UP|Q9FVC3 (Q9FVC3) Apyrase GS50 (Fragment), complete 461 e-130
TC217860 similar to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (56%) 217 7e-57
TC217859 weakly similar to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, part... 191 5e-49
TC232404 similar to UP|Q84UD8 (Q84UD8) Apyrase-like protein, par... 181 7e-46
TC224188 homologue to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (... 169 3e-42
TC221758 homologue to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (... 164 7e-41
TC206480 homologue to UP|Q84UD8 (Q84UD8) Apyrase-like protein, p... 164 7e-41
AW306606 similar to GP|11225135|gb apyrase GS50 {Glycine soja}, ... 149 2e-36
BI969027 homologue to GP|11225137|gb apyrase GS52 {Glycine soja}... 107 1e-23
BQ094726 similar to GP|19352177|db PsAPY2 {Pisum sativum}, parti... 93 2e-19
BE440256 weakly similar to GP|27804684|gb| apyrase-like protein ... 72 3e-13
BE821103 GP|11225135|gb apyrase GS50 {Glycine soja}, partial (23%) 67 1e-11
TC206481 similar to UP|Q84UD8 (Q84UD8) Apyrase-like protein, par... 65 4e-11
AI442610 37 0.016
TC215226 similar to UP|Q9SPM7 (Q9SPM7) Apyrase, partial (15%) 35 0.047
BF009816 32 0.52
BU965231 31 0.88
>TC222676 UP|Q84L88 (Q84L88) Apyrase, complete
Length = 1368
Score = 710 bits (1832), Expect = 0.0
Identities = 354/409 (86%), Positives = 376/409 (91%)
Frame = +1
Query: 1 MEFLITLITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHV 60
ME LI LIT LL MPAITSSQYLGNNLLT+RKIF K ISSYAVVFDAGSTGSRIHV
Sbjct: 1 MELLIKLIT-FLLFSMPAITSSQYLGNNLLTSRKIFLKHVEISSYAVVFDAGSTGSRIHV 177
Query: 61 YHFDQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTP 120
YHF+QNLDLLHIGK VE++NKITPGLSSYAN+PEQAAKSLIPLL+QAE+VVP DL KTP
Sbjct: 178 YHFNQNLDLLHIGKGVEYYNKITPGLSSYANNPEQAAKSLIPLLEQAEDVVPDDLQPKTP 357
Query: 121 IRLGATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYA 180
+RLGATAGLRLLNGDASEKILQ+VRDM SNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYA
Sbjct: 358 VRLGATAGLRLLNGDASEKILQSVRDMLSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYA 537
Query: 181 LGNLGKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLY 240
LGNLGKKYTKTVGV+DLGGGSVQMAYAVSKKTAKNAPKVADG DPYIKK+VLKG PYDLY
Sbjct: 538 LGNLGKKYTKTVGVIDLGGGSVQMAYAVSKKTAKNAPKVADGDDPYIKKVVLKGIPYDLY 717
Query: 241 VHSYLHFGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKI 300
VHSYLHFGREASRAEI+K+T SPNPCLLAGF+G YTY+GEEFKA A SGANFN CK
Sbjct: 718 VHSYLHFGREASRAEILKLTPRSPNPCLLAGFNGIYTYSGEEFKATAYTSGANFNKCKNT 897
Query: 301 IRKALKLNYPCPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRP 360
IRKALKLNYPCPYQNCTFGGIWNGGGGNGQK+ FASSSFFYLPED GMVD TPNF +RP
Sbjct: 898 IRKALKLNYPCPYQNCTFGGIWNGGGGNGQKNPFASSSFFYLPEDTGMVDASTPNFILRP 1077
Query: 361 VDLVSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
VD+ ++AK+ACALNFEDAKSTYPFL KKN+ASYVCMDLIYQYVLLVDGF
Sbjct: 1078VDIETKAKEACALNFEDAKSTYPFLDKKNVASYVCMDLIYQYVLLVDGF 1224
>TC212145 UP|Q76IB9 (Q76IB9) Apyrase (Fragment), complete
Length = 1173
Score = 301 bits (770), Expect(3) = e-139
Identities = 144/180 (80%), Positives = 157/180 (87%), Gaps = 2/180 (1%)
Frame = +1
Query: 232 LKGKPYDLYV--HSYLHFGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPA 289
L + Y + V +SYLHFGREASRAEI+K+T SPNPCLLAG +G YTY+GEEFKA A
Sbjct: 490 LYSREYHMIVMFNSYLHFGREASRAEILKLTPRSPNPCLLAGLNGIYTYSGEEFKATAYT 669
Query: 290 SGANFNGCKKIIRKALKLNYPCPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMV 349
SGANFN CK IRKALKLNYPCPYQNCTFGGIWNGGGGNGQK+LFASSSFFYLPED GMV
Sbjct: 670 SGANFNKCKNTIRKALKLNYPCPYQNCTFGGIWNGGGGNGQKNLFASSSFFYLPEDTGMV 849
Query: 350 DPKTPNFKIRPVDLVSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
D TPNF +RPVD+ ++AKKACALNFEDAKSTYPFL KKN+ASYVCMDLIYQYVLLVDGF
Sbjct: 850 DASTPNFILRPVDIETKAKKACALNFEDAKSTYPFLDKKNVASYVCMDLIYQYVLLVDGF 1029
Score = 201 bits (511), Expect(3) = e-139
Identities = 104/136 (76%), Positives = 113/136 (82%)
Frame = +2
Query: 65 QNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTPIRLG 124
+ LDLLHIGK VE++NKITPGLSSYAN+PEQAAKSLIPLL+QAE+VVP DL KTP+RLG
Sbjct: 38 RTLDLLHIGKGVEYYNKITPGLSSYANNPEQAAKSLIPLLEQAEDVVPDDLQPKTPVRLG 217
Query: 125 ATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGNL 184
ILQ+ RDM SNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYA GNL
Sbjct: 218 ---------------ILQSGRDMLSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYAWGNL 352
Query: 185 GKKYTKTVGVMDLGGG 200
GKKYTKTVGV+DLGGG
Sbjct: 353 GKKYTKTVGVIDLGGG 400
Score = 75.1 bits (183), Expect = 5e-14
Identities = 38/46 (82%), Positives = 39/46 (84%)
Frame = +3
Query: 200 GSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLYVHSYL 245
GSVQMA AVSKKTAKNAPKVADG DPYIKK+VLKG PYD YV L
Sbjct: 399 GSVQMADAVSKKTAKNAPKVADGDDPYIKKVVLKGIPYDRYVQQLL 536
Score = 34.3 bits (77), Expect(3) = e-139
Identities = 14/15 (93%), Positives = 15/15 (99%)
Frame = +1
Query: 53 STGSRIHVYHFDQNL 67
STGSRIHVYHF+QNL
Sbjct: 1 STGSRIHVYHFNQNL 45
>TC215225 UP|Q9FVC3 (Q9FVC3) Apyrase GS50 (Fragment), complete
Length = 1786
Score = 461 bits (1185), Expect = e-130
Identities = 231/411 (56%), Positives = 300/411 (72%), Gaps = 5/411 (1%)
Frame = +3
Query: 4 LITLITTVLLLLMPAITSSQ-YLGNNLLTNRKI---FQKQETIS-SYAVVFDAGSTGSRI 58
L L++TVL + +SS YLG ++RK+ + +TI SYAV+FDAGSTGSR+
Sbjct: 45 LSLLLSTVLATASSSSSSSSLYLGKGF-SHRKLSPSYHIHKTIDESYAVIFDAGSTGSRV 221
Query: 59 HVYHFDQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHK 118
HVY F+Q LDLL IG+D+E F K PGLS+YA +P+ AA+SLIPLL++AE VP + H +
Sbjct: 222 HVYRFNQQLDLLRIGQDLELFVKTMPGLSAYAENPQDAAESLIPLLEEAEAAVPQEFHPR 401
Query: 119 TPIRLGATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVN 178
TP++LGATAGLR L GDAS++ILQAV DM NRST NV DAVS++ G QEG+Y WVT+N
Sbjct: 402 TPVKLGATAGLRQLEGDASDRILQAVSDMLKNRSTLNVGADAVSVLSGNQEGAYQWVTIN 581
Query: 179 YALGNLGKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYD 238
Y LGNLGK Y++TV V+DLGGGSVQMAYAVS+ A AP+ DGV+ YI ++ L+GK Y
Sbjct: 582 YLLGNLGKHYSETVAVVDLGGGSVQMAYAVSETDAAKAPRAPDGVESYITEMFLRGKKYY 761
Query: 239 LYVHSYLHFGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCK 298
LYVHSYL +G A+RAE +KV R S NPC+LAGFDG Y Y G ++KAKAP SG++F+ C+
Sbjct: 762 LYVHSYLRYGMLAARAEALKV-RDSENPCILAGFDGYYVYGGVQYKAKAPPSGSSFSQCQ 938
Query: 299 KIIRKALKLNYPCPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKI 358
++ +AL +N C Y++CTFGGIWNGGGG G+ + F +S FF + ++ G VDP PN K+
Sbjct: 939 NVVVEALHVNATCSYKDCTFGGIWNGGGGAGENNFFIASFFFEVADEAGFVDPNAPNAKV 1118
Query: 359 RPVDLVSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
RPVD + AK AC +D KS +P K Y+C+DL+Y+Y LLVDGF
Sbjct: 1119RPVDFENAAKVACNTELKDLKSIFP-RVKDGDVPYICLDLVYEYTLLVDGF 1268
>TC217860 similar to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (56%)
Length = 881
Score = 217 bits (553), Expect = 7e-57
Identities = 121/195 (62%), Positives = 148/195 (75%), Gaps = 2/195 (1%)
Frame = +3
Query: 1 MEFLITLITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETI-SSYAVVFDAGSTGSRIH 59
M FL TVLL ++PA +SSQYLGNNLLT+RKIF K++ I +SYAV+FD GSTG+R+H
Sbjct: 6 MNFLT--FVTVLLFILPATSSSQYLGNNLLTHRKIFLKKDNIITSYAVIFDGGSTGTRVH 179
Query: 60 VYHFDQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKT 119
V+HFDQNLDLL IG +E KITPGLS+Y +DPEQAA+SLIPLL++AE+VVP DL T
Sbjct: 180 VFHFDQNLDLLPIGDSLELNKKITPGLSAYEDDPEQAAESLIPLLEEAESVVPEDLRPNT 359
Query: 120 PIRLGATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWV-TVN 178
P+RLGATAGLRLL G+ASE+ILQAVRDM SNRST N+Q DA S + S L+V +
Sbjct: 360 PVRLGATAGLRLLKGNASEQILQAVRDMLSNRSTLNLQSDA-SNYS*WKPRSCLYVGGIE 536
Query: 179 YALGNLGKKYTKTVG 193
++G GK K G
Sbjct: 537 LSIGQFGKSDFKDGG 581
Score = 196 bits (499), Expect = 1e-50
Identities = 94/140 (67%), Positives = 115/140 (82%)
Frame = +1
Query: 153 TFNVQPDAVSIIDGTQEGSYLWVTVNYALGNLGKKYTKTVGVMDLGGGSVQMAYAVSKKT 212
T+N+ P V+I+DG QE +Y+WV +NY LGNLGK +KTVGV DLGGGSVQMAYAVSK T
Sbjct: 466 TYNLMP--VTILDGNQEAAYMWVALNYLLGNLGKVISKTVGVADLGGGSVQMAYAVSKNT 639
Query: 213 AKNAPKVADGVDPYIKKLVLKGKPYDLYVHSYLHFGREASRAEIMKVTRSSPNPCLLAGF 272
AKNAP+ +G + YIK LVL GK YDLYVHSYLHFG+EASRAE++KVT S NPC+LAG+
Sbjct: 640 AKNAPQPPEGEESYIKTLVLNGKTYDLYVHSYLHFGKEASRAEMLKVTGDSANPCILAGY 819
Query: 273 DGTYTYAGEEFKAKAPASGA 292
+GTYTY+G ++KA A SG+
Sbjct: 820 NGTYTYSGVKYKALASTSGS 879
>TC217859 weakly similar to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (41%)
Length = 685
Score = 191 bits (485), Expect = 5e-49
Identities = 85/156 (54%), Positives = 115/156 (73%)
Frame = +2
Query: 254 AEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKIIRKALKLNYPCPY 313
AE+ VT SPNPC++A +DGTYTY+G E+KA A G+NF+ C+++ KALK+N PCP+
Sbjct: 2 AEMSNVTGDSPNPCIVAAYDGTYTYSGVEYKALASTCGSNFDKCREVALKALKVNEPCPH 181
Query: 314 QNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRPVDLVSEAKKACAL 373
QNCTFGGIWNGGGG+GQK L+ ++SF+YL G+ D + K+ P + + AK+AC +
Sbjct: 182 QNCTFGGIWNGGGGSGQKVLYVTTSFYYLVIQAGIADASKTSSKVYPAEFKAAAKRACQV 361
Query: 374 NFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
FEDA+STYP L ++ Y+CMD+ YQY LLVDGF
Sbjct: 362 KFEDAQSTYP-LMMEDALPYICMDITYQYTLLVDGF 466
>TC232404 similar to UP|Q84UD8 (Q84UD8) Apyrase-like protein, partial (28%)
Length = 535
Score = 181 bits (458), Expect = 7e-46
Identities = 80/132 (60%), Positives = 108/132 (81%)
Frame = +1
Query: 146 DMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGNLGKKYTKTVGVMDLGGGSVQMA 205
D+ +RS+ + DAV+++DGTQEG+Y WVT+NY LGNLGK Y+KTVGV+DLGGGSVQMA
Sbjct: 1 DLLKDRSSLKSESDAVTVLDGTQEGAYQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMA 180
Query: 206 YAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLYVHSYLHFGREASRAEIMKVTRSSPN 265
YA+S+ A APKV DG DPY+K++ L+G+ Y LYVHSYLH+G A+RAEI+K++ + N
Sbjct: 181 YAISETDAAMAPKVVDGGDPYLKEMFLRGRKYYLYVHSYLHYGLLAARAEILKISDDAEN 360
Query: 266 PCLLAGFDGTYT 277
PC+++G+DG YT
Sbjct: 361 PCVISGYDGKYT 396
>TC224188 homologue to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (28%)
Length = 407
Score = 169 bits (427), Expect = 3e-42
Identities = 81/127 (63%), Positives = 108/127 (84%), Gaps = 1/127 (0%)
Frame = +3
Query: 1 MEFLITLITTVLLLLMPAITSSQYL-GNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIH 59
M+FL +LL + A++S+QY GN LLT+RKIF KQE I+SYAV+FDAGSTGSR+H
Sbjct: 27 MDFLTLFTLLLLLFIHTALSSTQYHDGNILLTHRKIFPKQEAITSYAVIFDAGSTGSRVH 206
Query: 60 VYHFDQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKT 119
V+HFDQNLDLLHIG ++EF++K+TPGLS+Y ++P+QAA+SLIPLL++AE+VVP DL+ T
Sbjct: 207 VFHFDQNLDLLHIGNELEFYDKVTPGLSAYTDNPQQAAESLIPLLEEAESVVPEDLYPTT 386
Query: 120 PIRLGAT 126
P++LGAT
Sbjct: 387 PVKLGAT 407
>TC221758 homologue to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (37%)
Length = 625
Score = 164 bits (415), Expect = 7e-41
Identities = 77/143 (53%), Positives = 99/143 (68%)
Frame = +1
Query: 267 CLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKIIRKALKLNYPCPYQNCTFGGIWNGGG 326
C+ AGFDG YTY E + P + C++++ +ALKLN CP+Q CTFGGIW+GG
Sbjct: 13 CI*AGFDGDYTYQREIIRLSLPFLAPVKDECREVVLQALKLNKSCPHQYCTFGGIWDGGR 192
Query: 327 GNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRPVDLVSEAKKACALNFEDAKSTYPFLA 386
G+GQK LF +SSF+YLP ++G++D PN KI PVDL EAK+AC EDAKSTYP LA
Sbjct: 193 GSGQKILFGTSSFYYLPTEIGIIDLNKPNSKIHPVDLEIEAKRACETKLEDAKSTYPNLA 372
Query: 387 KKNIASYVCMDLIYQYVLLVDGF 409
+ + YVC+D+ YQY L DGF
Sbjct: 373 EDRL-PYVCLDIAYQYALYTDGF 438
>TC206480 homologue to UP|Q84UD8 (Q84UD8) Apyrase-like protein, partial (39%)
Length = 906
Score = 164 bits (415), Expect = 7e-41
Identities = 76/146 (52%), Positives = 101/146 (69%), Gaps = 1/146 (0%)
Frame = +1
Query: 265 NPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKIIRKALKLNYP-CPYQNCTFGGIWN 323
NPC+++G+DG+Y Y G+ FKA + +SGA+ N CK + +ALK+N C + CTFGGIWN
Sbjct: 13 NPCIISGYDGSYNYGGKSFKASSGSSGASLNECKSVALRALKVNESTCTHMKCTFGGIWN 192
Query: 324 GGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRPVDLVSEAKKACALNFEDAKSTYP 383
GGGG+GQK+LF +S FF + G DP P +RP D AK+AC E+AKST+P
Sbjct: 193 GGGGDGQKNLFVASFFFDRAAEAGFADPNLPVAIVRPADFEDAAKQACQTKLENAKSTFP 372
Query: 384 FLAKKNIASYVCMDLIYQYVLLVDGF 409
+ + N+ Y+CMDLIYQY LLVDGF
Sbjct: 373 RVDEGNL-PYLCMDLIYQYTLLVDGF 447
>AW306606 similar to GP|11225135|gb apyrase GS50 {Glycine soja}, partial
(33%)
Length = 467
Score = 149 bits (376), Expect = 2e-36
Identities = 76/131 (58%), Positives = 96/131 (73%)
Frame = +3
Query: 44 SYAVVFDAGSTGSRIHVYHFDQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPL 103
SYAV+FDAGSTGSR+HVY F+Q LDLL IG+ +E K PGLS+YA +P+ AA SLI L
Sbjct: 75 SYAVIFDAGSTGSRVHVYRFNQQLDLLLIGQYLELLVKTMPGLSAYAENPQDAA*SLISL 254
Query: 104 LQQAENVVPIDLHHKTPIRLGATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSI 163
L++A VP +LH TP++L ATA L+ L DAS+KILQAV DM NRST NV+ DA ++
Sbjct: 255 LEEA*AAVPQELHPMTPVKLVATARLKHLKSDASDKILQAVNDMLKNRSTLNVEADADAV 434
Query: 164 IDGTQEGSYLW 174
+ QE +Y W
Sbjct: 435 LSANQERAYQW 467
>BI969027 homologue to GP|11225137|gb apyrase GS52 {Glycine soja}, partial
(29%)
Length = 423
Score = 107 bits (266), Expect = 1e-23
Identities = 49/87 (56%), Positives = 62/87 (70%)
Frame = -1
Query: 312 PYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRPVDLVSEAKKAC 371
P+QNC FGGIW+GG G+GQK LF +SSF+YLP +G++D PN KI PVDL E K+AC
Sbjct: 423 PHQNCAFGGIWDGGRGSGQKILFGTSSFYYLPTXIGIIDLNKPNCKIHPVDLEIEVKRAC 244
Query: 372 ALNFEDAKSTYPFLAKKNIASYVCMDL 398
E AKSTYP L + + YVC+D+
Sbjct: 243 GSKLEXAKSTYPNLDEDRL-PYVCLDI 166
>BQ094726 similar to GP|19352177|db PsAPY2 {Pisum sativum}, partial (22%)
Length = 421
Score = 93.2 bits (230), Expect = 2e-19
Identities = 40/76 (52%), Positives = 62/76 (80%)
Frame = +1
Query: 7 LITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHVYHFDQN 66
L+ T +L +MP+ TS++ +G+ L NRK+ +++++ SYAV+FDAGS+GSR+HV+HFD N
Sbjct: 124 LLITFVLYVMPSSTSNESVGDYALVNRKMSPEKKSVGSYAVIFDAGSSGSRVHVFHFDHN 303
Query: 67 LDLLHIGKDVEFFNKI 82
LDL+HIGKD+E F ++
Sbjct: 304 LDLVHIGKDLELFVQV 351
>BE440256 weakly similar to GP|27804684|gb| apyrase-like protein {Medicago
truncatula}, partial (20%)
Length = 298
Score = 72.4 bits (176), Expect = 3e-13
Identities = 38/99 (38%), Positives = 51/99 (51%)
Frame = +2
Query: 311 CPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRPVDLVSEAKKA 370
C + CTF GIWNGGGG+G K+LF +S FF + DP P P ++ A
Sbjct: 5 CTHMKCTFRGIWNGGGGDGHKNLFVASLFFDRSSEACSTDPNLPVVIYHPANIYDTLTHA 184
Query: 371 CALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
C E+A T P + N+ Y +DLIY + LL D +
Sbjct: 185 CQT*LENA*FTLPLVHDLNLI-YHRIDLIYHHTLLQDPY 298
>BE821103 GP|11225135|gb apyrase GS50 {Glycine soja}, partial (23%)
Length = 501
Score = 67.4 bits (163), Expect = 1e-11
Identities = 31/61 (50%), Positives = 39/61 (63%)
Frame = -2
Query: 349 VDPKTPNFKIRPVDLVSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDG 408
VDP PN K+RPVD + AK AC +D KS +P K Y+C+DL+Y+Y LLVDG
Sbjct: 500 VDPNAPNAKVRPVDFENAAKVACNTELKDLKSIFP-RVKDGDVPYICLDLVYEYTLLVDG 324
Query: 409 F 409
F
Sbjct: 323 F 321
>TC206481 similar to UP|Q84UD8 (Q84UD8) Apyrase-like protein, partial (11%)
Length = 315
Score = 65.5 bits (158), Expect = 4e-11
Identities = 30/50 (60%), Positives = 36/50 (72%), Gaps = 1/50 (2%)
Frame = +2
Query: 292 ANFNGCKKIIRKALKLNYP-CPYQNCTFGGIWNGGGGNGQKHLFASSSFF 340
A+ N CK I +ALK+N C + CTFGGIWNGGGG+GQK+LF S FF
Sbjct: 2 ASLNECKNIAFQALKVNESKCTHMKCTFGGIWNGGGGDGQKNLFVDSFFF 151
>AI442610
Length = 253
Score = 37.0 bits (84), Expect = 0.016
Identities = 19/56 (33%), Positives = 28/56 (49%)
Frame = +2
Query: 354 PNFKIRPVDLVSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
P +R AC + AK +P +A + +Y+ M+LIYQ+ LL DGF
Sbjct: 8 PALIVRTAHFEDATNPACLSKLDSAKYIFPRVADGYL-NYISMELIYQHTLLADGF 172
>TC215226 similar to UP|Q9SPM7 (Q9SPM7) Apyrase, partial (15%)
Length = 331
Score = 35.4 bits (80), Expect = 0.047
Identities = 12/17 (70%), Positives = 16/17 (93%)
Frame = +1
Query: 393 YVCMDLIYQYVLLVDGF 409
Y+C+DL+Y+Y LLVDGF
Sbjct: 37 YICLDLVYEYTLLVDGF 87
>BF009816
Length = 431
Score = 32.0 bits (71), Expect = 0.52
Identities = 27/110 (24%), Positives = 41/110 (36%), Gaps = 27/110 (24%)
Frame = +1
Query: 237 YDLYVHSYLHFGREASRAEIMKVTRSSP--------------NPCLLAGFDGTYTYAGEE 282
Y+LY HS+LHFG A++ + + +PC G Y+Y E
Sbjct: 70 YNLYSHSFLHFGLNAAQDSLKEALVLGEFNLASQSLQKGLRIDPCTPTG----YSYNVES 237
Query: 283 FKAKAPASG------------ANFNGCKKIIRKAL-KLNYPCPYQNCTFG 319
+K + NF+ C+ + L K C YQ+C G
Sbjct: 238 WKFPPSSESEKNQYQSTVQARGNFSECRSVALTLLQKGKESCSYQHCDIG 387
>BU965231
Length = 445
Score = 31.2 bits (69), Expect = 0.88
Identities = 13/40 (32%), Positives = 21/40 (52%)
Frame = +1
Query: 36 FQKQETISSYAVVFDAGSTGSRIHVYHFDQNLDLLHIGKD 75
F E Y ++ D GSTG+R+HV+ + L G++
Sbjct: 304 FSPPEANFRYRIIVDGGSTGTRVHVFKYRSGRALEFSGRE 423
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,128,731
Number of Sequences: 63676
Number of extensions: 234711
Number of successful extensions: 1281
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 1258
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1266
length of query: 409
length of database: 12,639,632
effective HSP length: 100
effective length of query: 309
effective length of database: 6,272,032
effective search space: 1938057888
effective search space used: 1938057888
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC123593.9


