

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123593.7 + phase: 0
(467 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
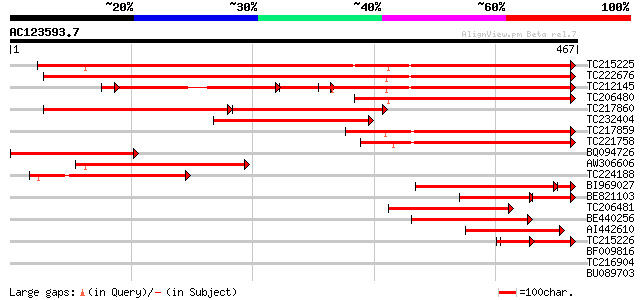
Score E
Sequences producing significant alignments: (bits) Value
TC215225 UP|Q9FVC3 (Q9FVC3) Apyrase GS50 (Fragment), complete 569 e-163
TC222676 UP|Q84L88 (Q84L88) Apyrase, complete 522 e-148
TC212145 UP|Q76IB9 (Q76IB9) Apyrase (Fragment), complete 233 e-103
TC206480 homologue to UP|Q84UD8 (Q84UD8) Apyrase-like protein, p... 353 9e-98
TC217860 similar to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (56%) 170 3e-86
TC232404 similar to UP|Q84UD8 (Q84UD8) Apyrase-like protein, par... 235 4e-62
TC217859 weakly similar to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, part... 226 2e-59
TC221758 homologue to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (... 195 4e-50
BQ094726 similar to GP|19352177|db PsAPY2 {Pisum sativum}, parti... 181 6e-46
AW306606 similar to GP|11225135|gb apyrase GS50 {Glycine soja}, ... 156 2e-38
TC224188 homologue to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (... 139 2e-33
BI969027 homologue to GP|11225137|gb apyrase GS52 {Glycine soja}... 123 1e-31
BE821103 GP|11225135|gb apyrase GS50 {Glycine soja}, partial (23%) 95 2e-31
TC206481 similar to UP|Q84UD8 (Q84UD8) Apyrase-like protein, par... 127 8e-30
BE440256 weakly similar to GP|27804684|gb| apyrase-like protein ... 126 2e-29
AI442610 99 5e-21
TC215226 similar to UP|Q9SPM7 (Q9SPM7) Apyrase, partial (15%) 56 4e-08
BF009816 37 0.025
TC216904 UP|P93166 (P93166) SCOF-1, complete 33 0.27
BU089703 homologue to GP|6002243|emb| Beta-glucosidase (EC 3.2.1... 30 2.3
>TC215225 UP|Q9FVC3 (Q9FVC3) Apyrase GS50 (Fragment), complete
Length = 1786
Score = 569 bits (1467), Expect = e-163
Identities = 289/449 (64%), Positives = 358/449 (79%), Gaps = 6/449 (1%)
Frame = +3
Query: 24 VVSIPILLITFVLYMMPSSSNYDSAGDYALLNRKLSPD----KKSGGSYAVIFDAGSSGS 79
++ + +LL T + SSS+ +RKLSP K SYAVIFDAGS+GS
Sbjct: 36 LIILSLLLSTVLATASSSSSSSSLYLGKGFSHRKLSPSYHIHKTIDESYAVIFDAGSTGS 215
Query: 80 RVHVFHFDQNLDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVPRELR 139
RVHV+ F+Q LDL+ IG+DLELF + PGLSAYA+ PQ AAESL+ LLE+AE VP+E
Sbjct: 216 RVHVYRFNQQLDLLRIGQDLELFVKTMPGLSAYAENPQDAAESLIPLLEEAEAAVPQEFH 395
Query: 140 SKTPVRIGATAGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQWVT 199
+TPV++GATAGLR LEGDASD+IL+AV D+LK+RS+ ADAV+VL G QEGAYQWVT
Sbjct: 396 PRTPVKLGATAGLRQLEGDASDRILQAVSDMLKNRSTLNVGADAVSVLSGNQEGAYQWVT 575
Query: 200 INYLLGNLGKDYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDGEDPYVKEMFLRGRK 259
INYLLGNLGK YS+TV VVDLGGGSVQMAYA+SE++AA AP+ DG + Y+ EMFLRG+K
Sbjct: 576 INYLLGNLGKHYSETVAVVDLGGGSVQMAYAVSETDAAKAPRAPDGVESYITEMFLRGKK 755
Query: 260 YYLYVHSYLRYGLLAARAEILKVAGDAENPCILSGSDGTYKYGGKSFKASS--SGASLNE 317
YYLYVHSYLRYG+LAARAE LKV D+ENPCIL+G DG Y YGG +KA + SG+S ++
Sbjct: 756 YYLYVHSYLRYGMLAARAEALKVR-DSENPCILAGFDGYYVYGGVQYKAKAPPSGSSFSQ 932
Query: 318 CKSVAHKALKVNESTCTHMKCTFGGIWNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPV 377
C++V +AL VN +TC++ CTFGGIWNGGGG G+ N F+ASFFF+ A EAGFVDPN+P
Sbjct: 933 CQNVVVEALHVN-ATCSYKDCTFGGIWNGGGGAGENNFFIASFFFEVADEAGFVDPNAPN 1109
Query: 378 AIVRPADFEDAAKQACQTKLENAKSTYPRVEEGNLPYLCMDLVYQYTLLVDGFGIYPWQE 437
A VRP DFE+AAK AC T+L++ KS +PRV++G++PY+C+DLVY+YTLLVDGFGI P QE
Sbjct: 1110AKVRPVDFENAAKVACNTELKDLKSIFPRVKDGDVPYICLDLVYEYTLLVDGFGIDPQQE 1289
Query: 438 ITLVKKVKYDDALVEAAWPLGSAIEAVSS 466
ITLV++V+Y D+LVEAAWPLGSAIEA+SS
Sbjct: 1290ITLVRQVEYQDSLVEAAWPLGSAIEAISS 1376
>TC222676 UP|Q84L88 (Q84L88) Apyrase, complete
Length = 1368
Score = 522 bits (1344), Expect = e-148
Identities = 254/441 (57%), Positives = 340/441 (76%), Gaps = 3/441 (0%)
Frame = +1
Query: 29 ILLITFVLYMMPSSSNYDSAGDYALLNRKLSPDKKSGGSYAVIFDAGSSGSRVHVFHFDQ 88
I LITF+L+ MP+ ++ G+ L +RK+ SYAV+FDAGS+GSR+HV+HF+Q
Sbjct: 13 IKLITFLLFSMPAITSSQYLGNNLLTSRKIFLKHVEISSYAVVFDAGSTGSRIHVYHFNQ 192
Query: 89 NLDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVPRELRSKTPVRIGA 148
NLDL+HIGK +E + ++ PGLS+YA P+QAA+SL+ LLE+AE VVP +L+ KTPVR+GA
Sbjct: 193 NLDLLHIGKGVEYYNKITPGLSSYANNPEQAAKSLIPLLEQAEDVVPDDLQPKTPVRLGA 372
Query: 149 TAGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQWVTINYLLGNLG 208
TAGLR L GDAS+KIL++VRD+L +RS+F DAV+++DGTQEG+Y WVT+NY LGNLG
Sbjct: 373 TAGLRLLNGDASEKILQSVRDMLSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGNLG 552
Query: 209 KDYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDGEDPYVKEMFLRGRKYYLYVHSYL 268
K Y+KTVGV+DLGGGSVQMAYA+S+ A AP+V DG+DPY+K++ L+G Y LYVHSYL
Sbjct: 553 KKYTKTVGVIDLGGGSVQMAYAVSKKTAKNAPKVADGDDPYIKKVVLKGIPYDLYVHSYL 732
Query: 269 RYGLLAARAEILKVAGDAENPCILSGSDGTYKYGGKSFKAS--SSGASLNECKSVAHKAL 326
+G A+RAEILK+ + NPC+L+G +G Y Y G+ FKA+ +SGA+ N+CK+ KAL
Sbjct: 733 HFGREASRAEILKLTPRSPNPCLLAGFNGIYTYSGEEFKATAYTSGANFNKCKNTIRKAL 912
Query: 327 KVNESTCTHMKCTFGGIWNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPADFE 386
K+N C + CTFGGIWNGGGG+GQKN F +S FF + G VD ++P I+RP D E
Sbjct: 913 KLN-YPCPYQNCTFGGIWNGGGGNGQKNPFASSSFFYLPEDTGMVDASTPNFILRPVDIE 1089
Query: 387 DAAKQACQTKLENAKSTYPRVEEGNL-PYLCMDLVYQYTLLVDGFGIYPWQEITLVKKVK 445
AK+AC E+AKSTYP +++ N+ Y+CMDL+YQY LLVDGFG+ P Q+IT K+++
Sbjct: 1090TKAKEACALNFEDAKSTYPFLDKKNVASYVCMDLIYQYVLLVDGFGLDPLQKITSGKEIE 1269
Query: 446 YDDALVEAAWPLGSAIEAVSS 466
Y DA+VEAAWPLG+A+EA+S+
Sbjct: 1270YQDAIVEAAWPLGNAVEAISA 1332
>TC212145 UP|Q76IB9 (Q76IB9) Apyrase (Fragment), complete
Length = 1173
Score = 233 bits (593), Expect(3) = e-103
Identities = 115/217 (52%), Positives = 159/217 (72%), Gaps = 5/217 (2%)
Frame = +1
Query: 255 LRGRKYYLYV--HSYLRYGLLAARAEILKVAGDAENPCILSGSDGTYKYGGKSFKAS--S 310
L R+Y++ V +SYL +G A+RAEILK+ + NPC+L+G +G Y Y G+ FKA+ +
Sbjct: 490 LYSREYHMIVMFNSYLHFGREASRAEILKLTPRSPNPCLLAGLNGIYTYSGEEFKATAYT 669
Query: 311 SGASLNECKSVAHKALKVNESTCTHMKCTFGGIWNGGGGDGQKNLFVASFFFDRAAEAGF 370
SGA+ N+CK+ KALK+N C + CTFGGIWNGGGG+GQKNLF +S FF + G
Sbjct: 670 SGANFNKCKNTIRKALKLN-YPCPYQNCTFGGIWNGGGGNGQKNLFASSSFFYLPEDTGM 846
Query: 371 VDPNSPVAIVRPADFEDAAKQACQTKLENAKSTYPRVEEGNL-PYLCMDLVYQYTLLVDG 429
VD ++P I+RP D E AK+AC E+AKSTYP +++ N+ Y+CMDL+YQY LLVDG
Sbjct: 847 VDASTPNFILRPVDIETKAKKACALNFEDAKSTYPFLDKKNVASYVCMDLIYQYVLLVDG 1026
Query: 430 FGIYPWQEITLVKKVKYDDALVEAAWPLGSAIEAVSS 466
FG+ P Q+IT K+++Y +A+VEAAWPLG+A+EA+S+
Sbjct: 1027FGLDPLQKITSGKEIEYQEAIVEAAWPLGNAVEAISA 1137
Score = 151 bits (382), Expect(3) = e-103
Identities = 74/136 (54%), Positives = 100/136 (73%)
Frame = +2
Query: 88 QNLDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVPRELRSKTPVRIG 147
+ LDL+HIGK +E + ++ PGLS+YA P+QAA+SL+ LLE+AE VVP +L+ KTPVR+G
Sbjct: 38 RTLDLLHIGKGVEYYNKITPGLSSYANNPEQAAKSLIPLLEQAEDVVPDDLQPKTPVRLG 217
Query: 148 ATAGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQWVTINYLLGNL 207
IL++ RD+L +RS+F DAV+++DGTQEG+Y WVT+NY GNL
Sbjct: 218 ---------------ILQSGRDMLSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYAWGNL 352
Query: 208 GKDYSKTVGVVDLGGG 223
GK Y+KTVGV+DLGGG
Sbjct: 353 GKKYTKTVGVIDLGGG 400
Score = 50.4 bits (119), Expect = 2e-06
Identities = 24/46 (52%), Positives = 32/46 (69%)
Frame = +3
Query: 223 GSVQMAYAISESEAAMAPQVMDGEDPYVKEMFLRGRKYYLYVHSYL 268
GSVQMA A+S+ A AP+V DG+DPY+K++ L+G Y YV L
Sbjct: 399 GSVQMADAVSKKTAKNAPKVADGDDPYIKKVVLKGIPYDRYVQQLL 536
Score = 30.8 bits (68), Expect(3) = e-103
Identities = 11/15 (73%), Positives = 15/15 (99%)
Frame = +1
Query: 76 SSGSRVHVFHFDQNL 90
S+GSR+HV+HF+QNL
Sbjct: 1 STGSRIHVYHFNQNL 45
>TC206480 homologue to UP|Q84UD8 (Q84UD8) Apyrase-like protein, partial (39%)
Length = 906
Score = 353 bits (906), Expect = 9e-98
Identities = 171/184 (92%), Positives = 177/184 (95%), Gaps = 2/184 (1%)
Frame = +1
Query: 285 DAENPCILSGSDGTYKYGGKSFKASS--SGASLNECKSVAHKALKVNESTCTHMKCTFGG 342
DAENPCI+SG DG+Y YGGKSFKASS SGASLNECKSVA +ALKVNESTCTHMKCTFGG
Sbjct: 4 DAENPCIISGYDGSYNYGGKSFKASSGSSGASLNECKSVALRALKVNESTCTHMKCTFGG 183
Query: 343 IWNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPADFEDAAKQACQTKLENAKS 402
IWNGGGGDGQKNLFVASFFFDRAAEAGF DPN PVAIVRPADFEDAAKQACQTKLENAKS
Sbjct: 184 IWNGGGGDGQKNLFVASFFFDRAAEAGFADPNLPVAIVRPADFEDAAKQACQTKLENAKS 363
Query: 403 TYPRVEEGNLPYLCMDLVYQYTLLVDGFGIYPWQEITLVKKVKYDDALVEAAWPLGSAIE 462
T+PRV+EGNLPYLCMDL+YQYTLLVDGFGIYPWQEITLVKKVKYDDALVEAAWPLGSAIE
Sbjct: 364 TFPRVDEGNLPYLCMDLIYQYTLLVDGFGIYPWQEITLVKKVKYDDALVEAAWPLGSAIE 543
Query: 463 AVSS 466
AVSS
Sbjct: 544 AVSS 555
>TC217860 similar to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (56%)
Length = 881
Score = 170 bits (430), Expect(2) = 3e-86
Identities = 81/128 (63%), Positives = 99/128 (77%)
Frame = +1
Query: 184 VTVLDGTQEGAYQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVM 243
VT+LDG QE AY WV +NYLLGNLGK SKTVGV DLGGGSVQMAYA+S++ A APQ
Sbjct: 484 VTILDGNQEAAYMWVALNYLLGNLGKVISKTVGVADLGGGSVQMAYAVSKNTAKNAPQPP 663
Query: 244 DGEDPYVKEMFLRGRKYYLYVHSYLRYGLLAARAEILKVAGDAENPCILSGSDGTYKYGG 303
+GE+ Y+K + L G+ Y LYVHSYL +G A+RAE+LKV GD+ NPCIL+G +GTY Y G
Sbjct: 664 EGEESYIKTLVLNGKTYDLYVHSYLHFGKEASRAEMLKVTGDSANPCILAGYNGTYTYSG 843
Query: 304 KSFKASSS 311
+KA +S
Sbjct: 844 VKYKALAS 867
Score = 167 bits (423), Expect(2) = 3e-86
Identities = 86/156 (55%), Positives = 119/156 (76%), Gaps = 1/156 (0%)
Frame = +3
Query: 29 ILLITFVLYMMPSSSNYDSAGDYALLNRKLSPDKKSG-GSYAVIFDAGSSGSRVHVFHFD 87
+ +T +L+++P++S+ G+ L +RK+ K + SYAVIFD GS+G+RVHVFHFD
Sbjct: 15 LTFVTVLLFILPATSSSQYLGNNLLTHRKIFLKKDNIITSYAVIFDGGSTGTRVHVFHFD 194
Query: 88 QNLDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVPRELRSKTPVRIG 147
QNLDL+ IG LEL +++ PGLSAY P+QAAESL+ LLE+AE VVP +LR TPVR+G
Sbjct: 195 QNLDLLPIGDSLELNKKITPGLSAYEDDPEQAAESLIPLLEEAESVVPEDLRPNTPVRLG 374
Query: 148 ATAGLRALEGDASDKILRAVRDLLKHRSSFKSDADA 183
ATAGLR L+G+AS++IL+AVRD+L +RS+ +DA
Sbjct: 375 ATAGLRLLKGNASEQILQAVRDMLSNRSTLNLQSDA 482
>TC232404 similar to UP|Q84UD8 (Q84UD8) Apyrase-like protein, partial (28%)
Length = 535
Score = 235 bits (599), Expect = 4e-62
Identities = 113/131 (86%), Positives = 124/131 (94%)
Frame = +1
Query: 169 DLLKHRSSFKSDADAVTVLDGTQEGAYQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMA 228
DLLK RSS KS++DAVTVLDGTQEGAYQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMA
Sbjct: 1 DLLKDRSSLKSESDAVTVLDGTQEGAYQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMA 180
Query: 229 YAISESEAAMAPQVMDGEDPYVKEMFLRGRKYYLYVHSYLRYGLLAARAEILKVAGDAEN 288
YAISE++AAMAP+V+DG DPY+KEMFLRGRKYYLYVHSYL YGLLAARAEILK++ DAEN
Sbjct: 181 YAISETDAAMAPKVVDGGDPYLKEMFLRGRKYYLYVHSYLHYGLLAARAEILKISDDAEN 360
Query: 289 PCILSGSDGTY 299
PC++SG DG Y
Sbjct: 361 PCVISGYDGKY 393
>TC217859 weakly similar to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (41%)
Length = 685
Score = 226 bits (576), Expect = 2e-59
Identities = 108/192 (56%), Positives = 140/192 (72%), Gaps = 2/192 (1%)
Frame = +2
Query: 277 AEILKVAGDAENPCILSGSDGTYKYGGKSFKA--SSSGASLNECKSVAHKALKVNESTCT 334
AE+ V GD+ NPCI++ DGTY Y G +KA S+ G++ ++C+ VA KALKVNE C
Sbjct: 2 AEMSNVTGDSPNPCIVAAYDGTYTYSGVEYKALASTCGSNFDKCREVALKALKVNEP-CP 178
Query: 335 HMKCTFGGIWNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPADFEDAAKQACQ 394
H CTFGGIWNGGGG GQK L+V + F+ +AG D + + V PA+F+ AAK+ACQ
Sbjct: 179 HQNCTFGGIWNGGGGSGQKVLYVTTSFYYLVIQAGIADASKTSSKVYPAEFKAAAKRACQ 358
Query: 395 TKLENAKSTYPRVEEGNLPYLCMDLVYQYTLLVDGFGIYPWQEITLVKKVKYDDALVEAA 454
K E+A+STYP + E LPY+CMD+ YQYTLLVDGFG+ PW+EI + +++Y ALVE A
Sbjct: 359 VKFEDAQSTYPLMMEDALPYICMDITYQYTLLVDGFGLDPWKEIIVANEIEYQGALVEGA 538
Query: 455 WPLGSAIEAVSS 466
WPLGSAIEA+SS
Sbjct: 539 WPLGSAIEAISS 574
>TC221758 homologue to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (37%)
Length = 625
Score = 195 bits (495), Expect = 4e-50
Identities = 95/179 (53%), Positives = 123/179 (68%), Gaps = 2/179 (1%)
Frame = +1
Query: 290 CILSGSDGTYKYGGKSFKASSSGAS--LNECKSVAHKALKVNESTCTHMKCTFGGIWNGG 347
CI +G DG Y Y + + S + +EC+ V +ALK+N+S C H CTFGGIW+GG
Sbjct: 13 CI*AGFDGDYTYQREIIRLSLPFLAPVKDECREVVLQALKLNKS-CPHQYCTFGGIWDGG 189
Query: 348 GGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPADFEDAAKQACQTKLENAKSTYPRV 407
G GQK LF S F+ E G +D N P + + P D E AK+AC+TKLE+AKSTYP +
Sbjct: 190 RGSGQKILFGTSSFYYLPTEIGIIDLNKPNSKIHPVDLEIEAKRACETKLEDAKSTYPNL 369
Query: 408 EEGNLPYLCMDLVYQYTLLVDGFGIYPWQEITLVKKVKYDDALVEAAWPLGSAIEAVSS 466
E LPY+C+D+ YQY L DGFG+ PWQEIT+ +++Y DALVEAAWPLG+AIEA+SS
Sbjct: 370 AEDRLPYVCLDIAYQYALYTDGFGLDPWQEITVANEIEYQDALVEAAWPLGTAIEAISS 546
>BQ094726 similar to GP|19352177|db PsAPY2 {Pisum sativum}, partial (22%)
Length = 421
Score = 181 bits (459), Expect = 6e-46
Identities = 89/106 (83%), Positives = 100/106 (93%)
Frame = +1
Query: 1 MLKRPTRQESLSDKIYRFRGTLLVVSIPILLITFVLYMMPSSSNYDSAGDYALLNRKLSP 60
MLKRP RQESL+DKIYRFRGTLLVV+IP+LLITFVLY+MPSS++ +S GDYAL+NRK+SP
Sbjct: 37 MLKRPGRQESLADKIYRFRGTLLVVAIPLLLITFVLYVMPSSTSNESVGDYALVNRKMSP 216
Query: 61 DKKSGGSYAVIFDAGSSGSRVHVFHFDQNLDLVHIGKDLELFEQLK 106
+KKS GSYAVIFDAGSSGSRVHVFHFD NLDLVHIGKDLELF Q+K
Sbjct: 217 EKKSVGSYAVIFDAGSSGSRVHVFHFDHNLDLVHIGKDLELFVQVK 354
>AW306606 similar to GP|11225135|gb apyrase GS50 {Glycine soja}, partial
(33%)
Length = 467
Score = 156 bits (394), Expect = 2e-38
Identities = 86/147 (58%), Positives = 106/147 (71%), Gaps = 4/147 (2%)
Frame = +3
Query: 55 NRKLSPD----KKSGGSYAVIFDAGSSGSRVHVFHFDQNLDLVHIGKDLELFEQLKPGLS 110
+ KLSP K SYAVIFDAGS+GSRVHV+ F+Q LDL+ IG+ LEL + PGLS
Sbjct: 27 HHKLSPSYHIHKTIDKSYAVIFDAGSTGSRVHVYRFNQQLDLLLIGQYLELLVKTMPGLS 206
Query: 111 AYAQKPQQAAESLVSLLEKAEGVVPRELRSKTPVRIGATAGLRALEGDASDKILRAVRDL 170
AYA+ PQ AA SL+SLLE+A VP+EL TPV++ ATA L+ L+ DASDKIL+AV D+
Sbjct: 207 AYAENPQDAA*SLISLLEEA*AAVPQELHPMTPVKLVATARLKHLKSDASDKILQAVNDM 386
Query: 171 LKHRSSFKSDADAVTVLDGTQEGAYQW 197
LK+RS+ +ADA VL QE AYQW
Sbjct: 387 LKNRSTLNVEADADAVLSANQERAYQW 467
>TC224188 homologue to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (28%)
Length = 407
Score = 139 bits (351), Expect = 2e-33
Identities = 73/136 (53%), Positives = 98/136 (71%), Gaps = 3/136 (2%)
Frame = +3
Query: 17 RFRGTL---LVVSIPILLITFVLYMMPSSSNYDSAGDYALLNRKLSPDKKSGGSYAVIFD 73
R RGT + +LL+ F+ + S+ +D G+ L +RK+ P +++ SYAVIFD
Sbjct: 6 RSRGTKNMDFLTLFTLLLLLFIHTALSSTQYHD--GNILLTHRKIFPKQEAITSYAVIFD 179
Query: 74 AGSSGSRVHVFHFDQNLDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGV 133
AGS+GSRVHVFHFDQNLDL+HIG +LE ++++ PGLSAY PQQAAESL+ LLE+AE V
Sbjct: 180 AGSTGSRVHVFHFDQNLDLLHIGNELEFYDKVTPGLSAYTDNPQQAAESLIPLLEEAESV 359
Query: 134 VPRELRSKTPVRIGAT 149
VP +L TPV++GAT
Sbjct: 360 VPEDLYPTTPVKLGAT 407
>BI969027 homologue to GP|11225137|gb apyrase GS52 {Glycine soja}, partial
(29%)
Length = 423
Score = 123 bits (308), Expect(2) = 1e-31
Identities = 57/119 (47%), Positives = 74/119 (61%), Gaps = 1/119 (0%)
Frame = -1
Query: 335 HMKCTFGGIWNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPADFEDAAKQACQ 394
H C FGGIW+GG G GQK LF S F+ G +D N P + P D E K+AC
Sbjct: 420 HQNCAFGGIWDGGRGSGQKILFGTSSFYYLPTXIGIIDLNKPNCKIHPVDLEIEVKRACG 241
Query: 395 TKLENAKSTYPRVEEGNLPYLCMDLVYQY-TLLVDGFGIYPWQEITLVKKVKYDDALVE 452
+KLE AKSTYP ++E LPY+C+D+ DGFG+ PWQEIT+ +++Y DALV+
Sbjct: 240 SKLEXAKSTYPNLDEDRLPYVCLDIAXXXCXCTXDGFGLDPWQEITVANEIEYQDALVK 64
Score = 31.6 bits (70), Expect(2) = 1e-31
Identities = 13/15 (86%), Positives = 15/15 (99%)
Frame = -3
Query: 452 EAAWPLGSAIEAVSS 466
EAAWPLG+AIEA+SS
Sbjct: 67 EAAWPLGTAIEAISS 23
>BE821103 GP|11225135|gb apyrase GS50 {Glycine soja}, partial (23%)
Length = 501
Score = 95.1 bits (235), Expect(2) = 2e-31
Identities = 41/62 (66%), Positives = 55/62 (88%)
Frame = -2
Query: 371 VDPNSPVAIVRPADFEDAAKQACQTKLENAKSTYPRVEEGNLPYLCMDLVYQYTLLVDGF 430
VDPN+P A VRP DFE+AAK AC T+L++ KS +PRV++G++PY+C+DLVY+YTLLVDGF
Sbjct: 500 VDPNAPNAKVRPVDFENAAKVACNTELKDLKSIFPRVKDGDVPYICLDLVYEYTLLVDGF 321
Query: 431 GI 432
G+
Sbjct: 320 GM 315
Score = 59.3 bits (142), Expect(2) = 2e-31
Identities = 28/36 (77%), Positives = 33/36 (90%)
Frame = -3
Query: 431 GIYPWQEITLVKKVKYDDALVEAAWPLGSAIEAVSS 466
GI P QEITLV++V+Y D+LVEAAWPLGSAIEA+SS
Sbjct: 193 GIDPQQEITLVRQVEYQDSLVEAAWPLGSAIEAISS 86
>TC206481 similar to UP|Q84UD8 (Q84UD8) Apyrase-like protein, partial (11%)
Length = 315
Score = 127 bits (320), Expect = 8e-30
Identities = 66/104 (63%), Positives = 75/104 (71%), Gaps = 1/104 (0%)
Frame = +2
Query: 313 ASLNECKSVAHKALKVNESTCTHMKCTFGGIWNGGGGDGQKNLFVASFFFDRAAEAGF-V 371
ASLNECK++A +ALKVNES CTHMKCTFGGIWNGGGGDGQKNLFV SFFFDR F +
Sbjct: 2 ASLNECKNIAFQALKVNESKCTHMKCTFGGIWNGGGGDGQKNLFVDSFFFDRGR*CWFCL 181
Query: 372 DPNSPVAIVRPADFEDAAKQACQTKLENAKSTYPRVEEGNLPYL 415
+ + P F +AC+TKL A STY RVE+GNL YL
Sbjct: 182 SKFTRCEGLVPWIFRLQLSKACKTKLAYATSTYQRVEDGNLSYL 313
>BE440256 weakly similar to GP|27804684|gb| apyrase-like protein {Medicago
truncatula}, partial (20%)
Length = 298
Score = 126 bits (317), Expect = 2e-29
Identities = 62/99 (62%), Positives = 70/99 (70%)
Frame = +2
Query: 332 TCTHMKCTFGGIWNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPADFEDAAKQ 391
TCTHMKCTF GIWNGGGGDG KNLFVAS FFDR++EA DPN PV I PA+ D
Sbjct: 2 TCTHMKCTFRGIWNGGGGDGHKNLFVASLFFDRSSEACSTDPNLPVVIYHPANIYDTLTH 181
Query: 392 ACQTKLENAKSTYPRVEEGNLPYLCMDLVYQYTLLVDGF 430
ACQT LENA T P V + NL Y +DL+Y +TLL D +
Sbjct: 182 ACQT*LENA*FTLPLVHDLNLIYHRIDLIYHHTLLQDPY 298
>AI442610
Length = 253
Score = 98.6 bits (244), Expect = 5e-21
Identities = 47/82 (57%), Positives = 58/82 (70%)
Frame = +2
Query: 376 PVAIVRPADFEDAAKQACQTKLENAKSTYPRVEEGNLPYLCMDLVYQYTLLVDGFGIYPW 435
P IVR A FEDA AC +KL++AK +PRV +G L Y+ M+L+YQ+TLL DGFG+Y W
Sbjct: 8 PALIVRTAHFEDATNPACLSKLDSAKYIFPRVADGYLNYISMELIYQHTLLADGFGLYTW 187
Query: 436 QEITLVKKVKYDDALVEAAWPL 457
+ IT V K K DD L EAAW L
Sbjct: 188 KYITSVIKSKNDDVLFEAAWSL 253
>TC215226 similar to UP|Q9SPM7 (Q9SPM7) Apyrase, partial (15%)
Length = 331
Score = 55.8 bits (133), Expect = 4e-08
Identities = 34/62 (54%), Positives = 40/62 (63%)
Frame = +2
Query: 405 PRVEEGNLPYLCMDLVYQYTLLVDGFGIYPWQEITLVKKVKYDDALVEAAWPLGSAIEAV 464
P + GNL L +L F I QEITLVK+V+Y D+LVEAAWPLGSAIEA+
Sbjct: 167 PNLYFGNLHLLKFNL----------FRIKSQQEITLVKQVEY*DSLVEAAWPLGSAIEAI 316
Query: 465 SS 466
SS
Sbjct: 317 SS 322
Score = 55.5 bits (132), Expect = 5e-08
Identities = 21/31 (67%), Positives = 30/31 (96%)
Frame = +1
Query: 402 STYPRVEEGNLPYLCMDLVYQYTLLVDGFGI 432
S +PRV++G++PY+C+DLVY+YTLLVDGFG+
Sbjct: 1 SIFPRVKDGDVPYICLDLVYEYTLLVDGFGM 93
>BF009816
Length = 431
Score = 36.6 bits (83), Expect = 0.025
Identities = 30/110 (27%), Positives = 47/110 (42%), Gaps = 28/110 (25%)
Frame = +1
Query: 260 YYLYVHSYLRYGLLAARAEILK--VAGDAE------------NPCILSGSDGTYKYGGKS 305
Y LY HS+L +GL AA+ + + V G+ +PC +G Y Y +S
Sbjct: 70 YNLYSHSFLHFGLNAAQDSLKEALVLGEFNLASQSLQKGLRIDPCTPTG----YSYNVES 237
Query: 306 FKASSSGAS--------------LNECKSVAHKALKVNESTCTHMKCTFG 341
+K S S +EC+SVA L+ + +C++ C G
Sbjct: 238 WKFPPSSESEKNQYQSTVQARGNFSECRSVALTLLQKGKESCSYQHCDIG 387
>TC216904 UP|P93166 (P93166) SCOF-1, complete
Length = 1114
Score = 33.1 bits (74), Expect = 0.27
Identities = 16/52 (30%), Positives = 27/52 (51%)
Frame = +2
Query: 308 ASSSGASLNECKSVAHKALKVNESTCTHMKCTFGGIWNGGGGDGQKNLFVAS 359
+S+SG +EC S+ HK+ ++ H +C + G NG + + VAS
Sbjct: 593 SSASGGKAHEC-SICHKSFPTGQALGGHKRCHYEGNGNGNNNNSNSVVTVAS 745
>BU089703 homologue to GP|6002243|emb| Beta-glucosidase (EC 3.2.1.21)
{Streptomyces coelicolor A3(2)}, partial (2%)
Length = 356
Score = 30.0 bits (66), Expect = 2.3
Identities = 18/80 (22%), Positives = 37/80 (45%), Gaps = 1/80 (1%)
Frame = +3
Query: 344 WNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAI-VRPADFEDAAKQACQTKLENAKS 402
+N G Q+++ + S P +P+ + +RP F ++ +C+ ++EN +
Sbjct: 105 FNSGSSSEQESVIIPS------------SPGTPLPLTIRPTCFSSSSSXSCEIEIENPNT 248
Query: 403 TYPRVEEGNLPYLCMDLVYQ 422
EEG L L + V++
Sbjct: 249 PASEEEEGLLKKLKSNEVFE 308
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,902,670
Number of Sequences: 63676
Number of extensions: 228459
Number of successful extensions: 1302
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 1262
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1286
length of query: 467
length of database: 12,639,632
effective HSP length: 101
effective length of query: 366
effective length of database: 6,208,356
effective search space: 2272258296
effective search space used: 2272258296
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC123593.7


