

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123593.3 + phase: 0 /pseudo
(176 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
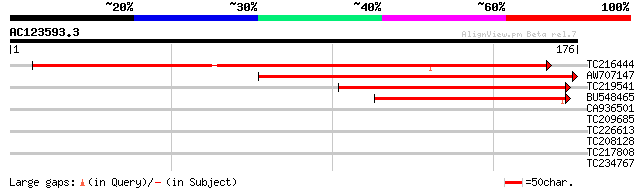
Score E
Sequences producing significant alignments: (bits) Value
TC216444 similar to UP|Q6L5N9 (Q6L5N9) Flowering locus T like pr... 150 3e-37
AW707147 similar to SP|Q9XH44|CET1_ CEN-like protein 1. [Common ... 119 6e-28
TC219541 similar to UP|Q84XL0 (Q84XL0) SP3D, partial (41%) 117 4e-27
BU548465 similar to GP|28200390|gb| SP3D {Lycopersicon esculentu... 79 1e-15
CA936501 30 0.59
TC209685 homologue to UP|Q61402 (Q61402) Gcap1 protein (Fragment... 29 1.3
TC226613 similar to UP|Q8H0V8 (Q8H0V8) Spliceosome associated pr... 27 3.8
TC208128 homologue to UP|Q8V7F3 (Q8V7F3) ORF1 (Fragment), partia... 27 5.0
TC217808 27 6.5
TC234767 similar to UP|Q94BT3 (Q94BT3) AT5g41810/K16L22_9, parti... 27 6.5
>TC216444 similar to UP|Q6L5N9 (Q6L5N9) Flowering locus T like protein,
complete
Length = 860
Score = 150 bits (379), Expect = 3e-37
Identities = 79/162 (48%), Positives = 107/162 (65%), Gaps = 1/162 (0%)
Frame = +2
Query: 8 NPLAVGRVIGDVLDPFESTIPLLVTYGNRTVTNGGELKPSQVANQPQVIIGVNDPTALYT 67
+PL VGRVIGDV+D F ++ + V +G++ VTNG ++KPS + P++ + N LYT
Sbjct: 104 DPLVVGRVIGDVVDMFIPSVNMSVYFGSKHVTNGCDIKPSIAISPPKLTLTGNMDN-LYT 280
Query: 68 LVLVDPDAPSPSYPSFREYLHWMVTDIPATNAASFGNEVVSYEKPRPNLGIHRFVFVLLH 127
LV+ DPDAPSPS PS RE++HW++ DIP G E+VSY PRP +GIHR++FVL
Sbjct: 281 LVMTDPDAPSPSEPSMREWIHWILVDIPGGTNPFRGKEIVSYVGPRPPIGIHRYIFVLFQ 460
Query: 128 QQ-CRQRVYAPGWRQNFNTREFIEFYNLGSPVAAVFFNCQRE 168
Q+ V P R +FNTR F +LG PVA V+FN Q+E
Sbjct: 461 QKGPLGLVEQPPTRASFNTRYFARQLDLGLPVATVYFNSQKE 586
>AW707147 similar to SP|Q9XH44|CET1_ CEN-like protein 1. [Common tobacco]
{Nicotiana tabacum}, partial (55%)
Length = 417
Score = 119 bits (299), Expect = 6e-28
Identities = 58/99 (58%), Positives = 68/99 (68%)
Frame = +3
Query: 78 PSYPSFREYLHWMVTDIPATNAASFGNEVVSYEKPRPNLGIHRFVFVLLHQQCRQRVYAP 137
PS P RE+LHWMVTDIP T SFG E+V YE P+P +GIHR+VF+L Q+ RQ V P
Sbjct: 3 PSDPCLREHLHWMVTDIPGTTDVSFGKEIVGYESPKPVIGIHRYVFILFKQRGRQTVRPP 182
Query: 138 GWRQNFNTREFIEFYNLGSPVAAVFFNCQRETGSGGRTF 176
R +FNTR F E LG PVAAV+FN QRET + R F
Sbjct: 183 SSRDHFNTRRFSEENGLGLPVAAVYFNAQRETAARRR*F 299
>TC219541 similar to UP|Q84XL0 (Q84XL0) SP3D, partial (41%)
Length = 758
Score = 117 bits (292), Expect = 4e-27
Identities = 54/72 (75%), Positives = 60/72 (83%)
Frame = +2
Query: 103 GNEVVSYEKPRPNLGIHRFVFVLLHQQCRQRVYAPGWRQNFNTREFIEFYNLGSPVAAVF 162
G+EVV+YE PRP +GIHR VFVL Q R+ VYAPGWRQNFNTREF E YNLG PVAAV+
Sbjct: 236 GHEVVTYESPRPMMGIHRLVFVLFRQLGRETVYAPGWRQNFNTREFAELYNLGLPVAAVY 415
Query: 163 FNCQRETGSGGR 174
FN QRE+GSGGR
Sbjct: 416 FNIQRESGSGGR 451
>BU548465 similar to GP|28200390|gb| SP3D {Lycopersicon esculentum}, partial
(29%)
Length = 694
Score = 79.0 bits (193), Expect = 1e-15
Identities = 38/62 (61%), Positives = 44/62 (70%), Gaps = 1/62 (1%)
Frame = -3
Query: 114 PNLGIHRFVFVLLHQQCRQRVYAPGWRQNFNTREFIEFYNLGSPVAAVFFNCQRETG-SG 172
P +GIHR V VL Q R + PGWRQNFNTR+F E YNLG PVAA++FNC+RE S
Sbjct: 692 PIVGIHRIVXVLFRQLRRLTLXXPGWRQNFNTRDFAEIYNLGLPVAAMYFNCKRENDQSS 513
Query: 173 GR 174
GR
Sbjct: 512 GR 507
>CA936501
Length = 266
Score = 30.0 bits (66), Expect = 0.59
Identities = 14/23 (60%), Positives = 16/23 (68%)
Frame = +2
Query: 154 LGSPVAAVFFNCQRETGSGGRTF 176
LG PVA V+FN QRET + R F
Sbjct: 2 LGLPVAVVYFNAQRETAARRR*F 70
>TC209685 homologue to UP|Q61402 (Q61402) Gcap1 protein (Fragment), partial
(20%)
Length = 1095
Score = 28.9 bits (63), Expect = 1.3
Identities = 13/44 (29%), Positives = 24/44 (54%)
Frame = -2
Query: 37 TVTNGGELKPSQVANQPQVIIGVNDPTALYTLVLVDPDAPSPSY 80
TV GG+++P + ++ +V+ G P LV D + P P++
Sbjct: 407 TVEGGGKMEPKMLVDEDEVVEGSLGPKR*SKLVNDDEEEPPPTF 276
>TC226613 similar to UP|Q8H0V8 (Q8H0V8) Spliceosome associated protein-like
(At4g21660), partial (59%)
Length = 1844
Score = 27.3 bits (59), Expect = 3.8
Identities = 20/75 (26%), Positives = 34/75 (44%), Gaps = 5/75 (6%)
Frame = +2
Query: 43 ELKPSQVANQPQVIIGVNDPTALYTLVLVDPDAPSPSYPSFREYLHWMVTDIPATNA--- 99
E+KP ++++ + +G+ + + L+ + P PSYP + IP NA
Sbjct: 551 EMKPGMLSHELKEALGMPEGSPPPWLINMQRYGPPPSYPHLK---------IPGLNAPIP 703
Query: 100 --ASFGNEVVSYEKP 112
ASFG + KP
Sbjct: 704 PGASFGYHPGGWGKP 748
>TC208128 homologue to UP|Q8V7F3 (Q8V7F3) ORF1 (Fragment), partial (19%)
Length = 711
Score = 26.9 bits (58), Expect = 5.0
Identities = 13/41 (31%), Positives = 19/41 (45%)
Frame = -1
Query: 44 LKPSQVANQPQVIIGVNDPTALYTLVLVDPDAPSPSYPSFR 84
LKP ++ P + + P A L + +PSP SFR
Sbjct: 513 LKPPPISRPPPTLTHKSPPVATQILQSIPSPSPSPPTTSFR 391
>TC217808
Length = 1106
Score = 26.6 bits (57), Expect = 6.5
Identities = 14/34 (41%), Positives = 22/34 (64%)
Frame = +3
Query: 66 YTLVLVDPDAPSPSYPSFREYLHWMVTDIPATNA 99
++L++VDP+ PS FRE L M ++PA+ A
Sbjct: 555 FSLLVVDPN-DEPSVNKFRECLKKMRAEVPASLA 653
>TC234767 similar to UP|Q94BT3 (Q94BT3) AT5g41810/K16L22_9, partial (23%)
Length = 745
Score = 26.6 bits (57), Expect = 6.5
Identities = 11/31 (35%), Positives = 20/31 (64%)
Frame = +1
Query: 60 NDPTALYTLVLVDPDAPSPSYPSFREYLHWM 90
N P +YTL +++ + SP +P+F+ L W+
Sbjct: 457 NMPMTVYTLFMINLNRFSPQFPNFQS-LRWV 546
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.139 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,146,006
Number of Sequences: 63676
Number of extensions: 119424
Number of successful extensions: 494
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 490
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 492
length of query: 176
length of database: 12,639,632
effective HSP length: 91
effective length of query: 85
effective length of database: 6,845,116
effective search space: 581834860
effective search space used: 581834860
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC123593.3


