

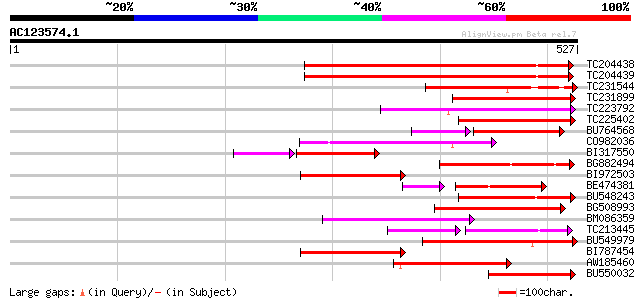
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123574.1 + phase: 0 /pseudo
(527 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 187 8e-48
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 186 2e-47
TC231544 weakly similar to UP|Q9FLR2 (Q9FLR2) Polyprotein-like, ... 157 8e-39
TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Frag... 153 2e-37
TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polypro... 127 1e-29
TC225402 weakly similar to UP|Q6I923 (Q6I923) Copia-like retrotr... 123 2e-28
BU764568 102 7e-28
CO982036 112 3e-25
BI317550 weakly similar to GP|9759590|dbj| polyprotein-like {Ara... 104 1e-23
BG882494 weakly similar to GP|27901694|gb| gag-pol polyprotein {... 107 1e-23
BI972503 weakly similar to GP|18378607|gb polyprotein {Oryza sat... 104 1e-22
BE474381 similar to GP|21434|emb|CA ORF4 {Solanum tuberosum}, pa... 88 2e-22
BU548243 103 2e-22
BG508993 101 7e-22
BM086359 100 2e-21
TC213445 70 4e-21
BU549979 97 1e-20
BI787454 weakly similar to GP|21434|emb|CA ORF4 {Solanum tuberos... 97 2e-20
AW185460 96 3e-20
BU550032 similar to GP|27901696|gb| gag-pol polyprotein {Vitis v... 95 9e-20
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 187 bits (476), Expect = 8e-48
Identities = 93/250 (37%), Positives = 152/250 (60%)
Frame = +1
Query: 275 LRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSDPSVK 334
+++ F++ +G+L YFLGLQV + I L Q KY +++ G+ ++ TP+ +K
Sbjct: 3847 MQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSKYAKNIVKKFGMENASHKRTPAPTHLK 4026
Query: 335 LHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMRVLKY 394
L +D + D S YR ++G LLYL +RPDIT+ +++ A H + R+LKY
Sbjct: 4027 LSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGVCARYQANPKISHLNQVKRILKY 4206
Query: 395 LKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVS 454
+ + G+ + S ++G+ DADWAG D R+S SG CF+LG +LISW +KKQ VS
Sbjct: 4207 VNGTSDYGIMYCHCSDSMLVGYCDADWAGSADDRKSTSGGCFYLGTNLISWFSKKQNCVS 4386
Query: 455 RSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFHERTKH 514
S+++ EY A ++ +L W+ +L++ ++ + LYCDN SA++I+ NP+ H RTKH
Sbjct: 4387 LSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVM-TLYCDNMSAINISKNPVQHSRTKH 4563
Query: 515 LEIDCHLVQD 524
++I H ++D
Sbjct: 4564 IDIRHHYIRD 4593
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 186 bits (473), Expect = 2e-47
Identities = 92/250 (36%), Positives = 152/250 (60%)
Frame = +1
Query: 275 LRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSDPSVK 334
+++ F++ +G+L YFLGLQV + I L Q +Y +++ G+ ++ TP+ +K
Sbjct: 3844 MQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSRYAKNIVKKFGMENASHKRTPAPTHLK 4023
Query: 335 LHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMRVLKY 394
L +D + D S YR ++G LLYL +RPDIT+ +++ A H + R+LKY
Sbjct: 4024 LSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGVCARYQANPKISHLTQVKRILKY 4203
Query: 395 LKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVS 454
+ + G+ + S ++G+ DADWAG D R+S SG CF+LG +LISW +KKQ VS
Sbjct: 4204 VNGTSDYGIMYCHCSNPMLVGYCDADWAGSADDRKSTSGGCFYLGNNLISWFSKKQNCVS 4383
Query: 455 RSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFHERTKH 514
S+++ EY A ++ +L W+ +L++ ++ + LYCDN SA++I+ NP+ H RTKH
Sbjct: 4384 LSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVM-TLYCDNMSAINISKNPVQHSRTKH 4560
Query: 515 LEIDCHLVQD 524
++I H ++D
Sbjct: 4561 IDIRHHYIRD 4590
>TC231544 weakly similar to UP|Q9FLR2 (Q9FLR2) Polyprotein-like, partial
(16%)
Length = 662
Score = 157 bits (398), Expect = 8e-39
Identities = 86/144 (59%), Positives = 103/144 (70%), Gaps = 3/144 (2%)
Frame = +3
Query: 387 AAMRVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWR 446
AA RVLKYLK KGL F R+S +QILGFSDADWA C+DS +SI+ CFFLG SLISW+
Sbjct: 18 AATRVLKYLKGCPRKGLSFSRESPIQILGFSDADWATCIDSSKSITWYCFFLGSSLISWK 197
Query: 447 TKKQLIVSRSSSKEE--YRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALH-IA 503
KKQ VSRSSS E YRAL + TCELQWL YLL+DL + ++YCDNQSAL +
Sbjct: 198 AKKQNTVSRSSSSSEAKYRALTSTTCELQWLTYLLKDLHVT-----LIYCDNQSALQ*LP 362
Query: 504 ANPIFHERTKHLEIDCHLVQDKLQ 527
I+H + LEIDCH+V++K Q
Sbjct: 363 IKVIYHGQ---LEIDCHIVREKTQ 425
>TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragment),
partial (30%)
Length = 687
Score = 153 bits (387), Expect = 2e-37
Identities = 66/115 (57%), Positives = 89/115 (77%)
Frame = +2
Query: 412 QILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSSKEEYRALAAATCE 471
Q+ G+ DADWAGC RRS SG C F+G +L+SW++KKQ +V+RSS++ EYR++A TCE
Sbjct: 14 QLSGYCDADWAGCPMDRRSTSGYCVFIGGNLVSWKSKKQTVVARSSAEAEYRSMAMVTCE 193
Query: 472 LQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFHERTKHLEIDCHLVQDKL 526
L W+ LQ+L+ LYCDNQ+ALHIA+NP+FHERTKH+EIDCH +++KL
Sbjct: 194 LMWIKQFLQELRFCEELQMKLYCDNQAALHIASNPVFHERTKHIEIDCHFIREKL 358
>TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polyprotein,
partial (7%)
Length = 804
Score = 127 bits (319), Expect = 1e-29
Identities = 61/185 (32%), Positives = 112/185 (59%), Gaps = 3/185 (1%)
Frame = +1
Query: 345 DISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMRVLKYLKTSLGKGLF 404
D++ +RRL+G L YL +RP+I F +S+F+ + H AA RVL+ +K ++G G+
Sbjct: 10 DVTEFRRLIGSLRYLCNSRPNICFAVSLISRFMKRPRLSHMQAAKRVLRLIKGTIGSGVL 189
Query: 405 FP---RDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSSKEE 461
FP + +LG++D+DW + +S G F + ++ +KKQ +++ S+ + E
Sbjct: 190 FPFKAKSGKPDLLGYTDSDWKRDPEQEKSTGGYLFMYNDAPVA*SSKKQDVIALSTCEAE 369
Query: 462 YRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFHERTKHLEIDCHL 521
Y A + C+ W++ LL++L+L+ K L DN+SA+++A +P H R+KH+E+ H
Sbjct: 370 YVAASLGACQAVWMMNLLEELKLRERKPVNLLIDNKSAINLAKHPTLHGRSKHIELRFHY 549
Query: 522 VQDKL 526
++D++
Sbjct: 550 IRDQV 564
>TC225402 weakly similar to UP|Q6I923 (Q6I923) Copia-like retrotransposon
Hopscotch polyprotein, partial (7%)
Length = 1446
Score = 123 bits (308), Expect = 2e-28
Identities = 55/109 (50%), Positives = 81/109 (73%)
Frame = +2
Query: 418 DADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSSKEEYRALAAATCELQWLLY 477
DA+WA R S G C +G++L+ W++ K +V+RSS++ EY+A+ ATCEL W+
Sbjct: 8 DANWAVSPIDRGSTLGYCVSIGENLVLWKSNK*NVVARSSAEAEYKAMTVATCELIWIKQ 187
Query: 478 LLQDLQLQTSKLHVLYCDNQSALHIAANPIFHERTKHLEIDCHLVQDKL 526
LLQ+L+ +++ L CDNQ+ALHIA+NP+FHERTKH+EIDCH V++K+
Sbjct: 188 LLQELKFGSTQQMKLCCDNQAALHIASNPVFHERTKHIEIDCHFVREKV 334
>BU764568
Length = 420
Score = 102 bits (254), Expect(2) = 7e-28
Identities = 47/84 (55%), Positives = 63/84 (74%)
Frame = +3
Query: 432 SGQCFFLGKSLISWRTKKQLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHV 491
SG C +G +LISW++KKQ +V++SS++ EYRA+A TCEL WL LL +L+ +
Sbjct: 168 SGYCVLIGGNLISWKSKKQSVVAKSSAEAEYRAMALVTCELIWLKQLL*ELKFEEDTQMT 347
Query: 492 LYCDNQSALHIAANPIFHERTKHL 515
L CDNQ+ALHIA+NPIFH RTKH+
Sbjct: 348 LICDNQAALHIASNPIFH*RTKHI 419
Score = 40.0 bits (92), Expect(2) = 7e-28
Identities = 23/55 (41%), Positives = 30/55 (53%)
Frame = +1
Query: 374 SQFLAKRTQVHHSAAMRVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSR 428
SQFL Q H +A +LK K++ GKGL + QI+G+SDAD G R
Sbjct: 1 SQFLNSPCQDHWNAVS*ILK*TKSAPGKGLIYEDKGHSQIIGYSDAD*VGSPSDR 165
>CO982036
Length = 674
Score = 112 bits (281), Expect = 3e-25
Identities = 68/186 (36%), Positives = 101/186 (53%), Gaps = 3/186 (1%)
Frame = -2
Query: 270 NIKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPS 329
N+ S L +SF +K LG+L YF+ ++V S + R ++ ++PIS+P
Sbjct: 592 NLTSKLNSSFPLKLLGKLDYFVEIEVK-SMPDLLFSLRTSIFEIFCRKPR*QAQPISSPM 416
Query: 330 DPSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAM 389
+ KL + S ++ + YR +VG L Y RP+I+F ++ QF++ H +
Sbjct: 415 TTTCKLSKSDSDLFSGPTFYRSVVGALQYTTVIRPEISFAVNKVCQFMSNPLDSHWTEVK 236
Query: 390 RVLKYLKTSLGKGL-FFPRDSA--LQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWR 446
R+L+YLK SL GL P S+ L I GF DADWA +D +RS SG FLG +LISW
Sbjct: 235 RILRYLKGSLSYGL*LKPAISSQPLPIRGFCDADWASAVDDKRSTSGAAVFLGPNLISWW 56
Query: 447 TKKQLI 452
KQ +
Sbjct: 55 XXKQQV 38
>BI317550 weakly similar to GP|9759590|dbj| polyprotein-like {Arabidopsis
thaliana}, partial (18%)
Length = 421
Score = 104 bits (259), Expect(2) = 1e-23
Identities = 50/77 (64%), Positives = 61/77 (78%)
Frame = -2
Query: 267 EFNNIKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPIS 326
EF+ IK+VL +FKIK+LG+LKYFLGL+VAHS+ GI++ QRKYCLDLL DSGL KP S
Sbjct: 234 EFDRIKNVLDLAFKIKNLGKLKYFLGLEVAHSRLGITISQRKYCLDLLKDSGLLGCKPAS 55
Query: 327 TPSDPSVKLHRDSSPPY 343
TP D S+KLH + PY
Sbjct: 54 TPLDTSIKLHSAAGTPY 4
Score = 24.3 bits (51), Expect(2) = 1e-23
Identities = 20/56 (35%), Positives = 27/56 (47%)
Frame = -3
Query: 209 MD*SEPPENGMKGSPLCYWLKDTSMLTHIIHYLPNLQLLHSPSFWSM*MILF*QEI 264
M *S+ +GMK +C+ K T I YL + + + S W M*M F Q I
Sbjct: 410 MG*SKQVGSGMKS*LICFSKKVTYNQFQITPYLLSPKEILSLHCWYM*MTSFLQVI 243
>BG882494 weakly similar to GP|27901694|gb| gag-pol polyprotein {Vitis
vinifera}, partial (20%)
Length = 400
Score = 107 bits (268), Expect = 1e-23
Identities = 56/126 (44%), Positives = 77/126 (60%)
Frame = -2
Query: 400 GKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSSK 459
GKGL + QI+G+ D DWAG +R C + LISW +KK +V+RS +K
Sbjct: 372 GKGLTYEDREHTQIVGYLDVDWAGSAIHKRPTCRYCVLIRGKLISWNSKKHNVVTRSRTK 193
Query: 460 EEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFHERTKHLEIDC 519
EYRA+ A C+L W L + LQ + + L CDNQ+AL+IA+N F RTKH+EI+C
Sbjct: 192 VEYRAM-ALICQLIWFKQLRKQLQFEDTMQMTLICDNQAALYIASNQ-FSTRTKHIEINC 19
Query: 520 HLVQDK 525
H V++K
Sbjct: 18 HFVREK 1
>BI972503 weakly similar to GP|18378607|gb polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (5%)
Length = 327
Score = 104 bits (259), Expect = 1e-22
Identities = 49/98 (50%), Positives = 69/98 (70%)
Frame = +2
Query: 271 IKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSD 330
+K L + F+ KDLG LKYFLG++VA S + + QRKY LD+L ++G+ + +P+ +P D
Sbjct: 32 LKEHLFSHFQTKDLGYLKYFLGIEVAQSGVDVVISQRKYALDILEETGMQNCRPVDSPMD 211
Query: 331 PSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITF 368
P++KL D S Y D YRRLVG+L+YL TRPDI+F
Sbjct: 212 PNLKLMADQSEIYHDPERYRRLVGKLIYLTITRPDISF 325
>BE474381 similar to GP|21434|emb|CA ORF4 {Solanum tuberosum}, partial (20%)
Length = 406
Score = 88.2 bits (217), Expect(2) = 2e-22
Identities = 44/85 (51%), Positives = 62/85 (72%)
Frame = +3
Query: 415 GFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSSKEEYRALAAATCELQW 474
G++DADWAG + RRS SG C F+G +L+S +KKQ +V+RSS++ E+RALA CE W
Sbjct: 153 GYTDADWAGSVTDRRSTSGYCTFVGGNLVS*-SKKQSVVARSSAEAEFRALAHGICETLW 329
Query: 475 LLYLLQDLQLQTSKLHVLYCDNQSA 499
+ LLQ+L++ +S LYCDN+SA
Sbjct: 330 VKKLLQELKVHSSPPIKLYCDNKSA 404
Score = 36.2 bits (82), Expect(2) = 2e-22
Identities = 17/39 (43%), Positives = 23/39 (58%)
Frame = +1
Query: 366 ITFVTQQLSQFLAKRTQVHHSAAMRVLKYLKTSLGKGLF 404
I F +SQF+ H AA R+L+YLK S G+GL+
Sbjct: 7 IAFAVSMVSQFMHAPGHEHLEAAFRILRYLKGSPGRGLY 123
>BU548243
Length = 599
Score = 103 bits (256), Expect = 2e-22
Identities = 50/109 (45%), Positives = 73/109 (66%)
Frame = -1
Query: 418 DADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSSKEEYRALAAATCELQWLLY 477
DA WA +D RS G FLG +LISW ++KQ + ++SS++ EYR++A + EL W+
Sbjct: 587 DAGWASDVDDHRSTLGSAIFLGPNLISWWSRKQQVTAQSSTEAEYRSIAQTSAELTWIQA 408
Query: 478 LLQDLQLQTSKLHVLYCDNQSALHIAANPIFHERTKHLEIDCHLVQDKL 526
LL +LQ+ + V+ CDN+SA+ IA N +FH RTKH+EID V +K+
Sbjct: 407 LLMELQIPFTP-PVILCDNKSAVAIAHNLVFHSRTKHMEIDVFFVHEKV 264
>BG508993
Length = 374
Score = 101 bits (252), Expect = 7e-22
Identities = 48/121 (39%), Positives = 78/121 (63%)
Frame = +1
Query: 396 KTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSR 455
K ++ GLF+ + +++GF D+D+AG +D R+S +G FF+G + +W +KKQ IV+
Sbjct: 4 KGTIDFGLFYSPSNNYKLVGFCDSDFAGDVDDRKSTTGFVFFMGDCVFTWSSKKQGIVTL 183
Query: 456 SSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFHERTKHL 515
+ + EY A + TC WL LL++LQL + +Y DN+SA +A N +FHER+KH+
Sbjct: 184 FTCEAEYVAATSCTCHAIWLRRLLEELQLLQKESTKIYVDNRSAQELAKNSVFHERSKHI 363
Query: 516 E 516
+
Sbjct: 364 D 366
>BM086359
Length = 427
Score = 100 bits (249), Expect = 2e-21
Identities = 57/142 (40%), Positives = 77/142 (54%)
Frame = +1
Query: 291 LGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSDPSVKLHRDSSPPYTDISAYR 350
LG+ VA S GI + Q KY LD+L ++G+ P +TP DP+VKL D
Sbjct: 1 LGIDVAQSSYGIVISQWKYALDILTETGMLDCLPSNTPMDPNVKLLSGQGEALEDPGR*C 180
Query: 351 RLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMRVLKYLKTSLGKGLFFPRDSA 410
LVGRL YL TR DITF LSQFL T +A +R+L+Y+K + G GL +
Sbjct: 181 CLVGRLNYLTVTRLDITFAVGVLSQFLKDPTDSQWNATIRILRYIKNAPGPGLLYEDKGN 360
Query: 411 LQILGFSDADWAGCLDSRRSIS 432
+++ + DADW G + S S
Sbjct: 361 GKVVCYFDADWPGSPSDKSSTS 426
>TC213445
Length = 705
Score = 70.1 bits (170), Expect(2) = 4e-21
Identities = 35/100 (35%), Positives = 58/100 (58%)
Frame = +1
Query: 424 CLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQ 483
C R S S C F+G +L+SW +KKQ V S+++ EY + + ++ W+ L D
Sbjct: 397 CKTDRESTSDTCHFIGSALVSWHSKKQNSVVLSTAEAEYISARSYYAQIFWMRQQLFDYG 576
Query: 484 LQTSKLHVLYCDNQSALHIAANPIFHERTKHLEIDCHLVQ 523
L+ + + CDN SA++++ N I + RTKH+EI H ++
Sbjct: 577 LKLDHIPI-RCDNTSAINLSKNHILYSRTKHIEIRHHFLR 693
Score = 49.7 bits (117), Expect(2) = 4e-21
Identities = 22/68 (32%), Positives = 41/68 (59%)
Frame = +2
Query: 352 LVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMRVLKYLKTSLGKGLFFPRDSAL 411
++ LYL+T+RP I F ++ A + H S R+++YL + GL++P++S+
Sbjct: 197 MIESFLYLSTSRPHIMFSVCMCVRYQANPKESHLSVIKRIMRYLLGIINLGLWYPKNSSY 376
Query: 412 QILGFSDA 419
++G+SDA
Sbjct: 377 NLVGYSDA 400
>BU549979
Length = 615
Score = 97.4 bits (241), Expect = 1e-20
Identities = 48/146 (32%), Positives = 89/146 (60%), Gaps = 2/146 (1%)
Frame = -1
Query: 384 HHSAAMRVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLI 443
H A +V++YL+ + L + + + L+++G+SD+D+AGC+DSRRS SG F L ++
Sbjct: 579 HWKTAKKVMRYLQGTKDYMLMYKQTNCLEVIGYSDSDFAGCVDSRRSTSGYIFMLADGVV 400
Query: 444 SWRTKKQLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQL--QTSKLHVLYCDNQSALH 501
SWR+ KQ +++ S+ + E+ AT WL + L++ S+ LYCDN +A+
Sbjct: 399 SWRSSKQTLIATSTMEVEFVPCFEATSHGVWLKSFMSSLRVVDSISRPLKLYCDNFAAVF 220
Query: 502 IAANPIFHERTKHLEIDCHLVQDKLQ 527
+A N R+KH++I +++++++
Sbjct: 219 MAKNNKSGNRSKHIDIKYLVIRERVK 142
>BI787454 weakly similar to GP|21434|emb|CA ORF4 {Solanum tuberosum}, partial
(21%)
Length = 421
Score = 97.1 bits (240), Expect = 2e-20
Identities = 47/98 (47%), Positives = 66/98 (66%)
Frame = +2
Query: 271 IKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSD 330
+K L F+ KDL LKYFLG++VA S G+ + QRKY LD+L ++G+ + + + +P D
Sbjct: 125 LKEHLFNHFQTKDLRYLKYFLGIEVAQSGDGVVISQRKYALDILEETGMQNCRLVDSPMD 304
Query: 331 PSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITF 368
P++KL S Y D YRRLVG+L+YL TRPDI+F
Sbjct: 305 PNLKLMAYQSEVYPDPERYRRLVGKLIYLTITRPDISF 418
>AW185460
Length = 411
Score = 96.3 bits (238), Expect = 3e-20
Identities = 44/112 (39%), Positives = 74/112 (65%), Gaps = 2/112 (1%)
Frame = +2
Query: 357 LYLNT--TRPDITFVTQQLSQFLAKRTQVHHSAAMRVLKYLKTSLGKGLFFPRDSALQIL 414
LY+ + TRPDI + T LS+F+ +Q+H A R+L+YL+ + G+++ ++ ++L
Sbjct: 71 LYIKSQATRPDIMYATSLLSRFMQSPSQIHFGAGKRILRYLQGTKAFGIWYTTETNSELL 250
Query: 415 GFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSSKEEYRALA 466
G++D+DWAG D +S SG F LG + SW +KKQ V++S+++ EY A+A
Sbjct: 251 GYTDSDWAGSTDDMKSTSGYAFSLGSGMFSWASKKQATVAQSTAEAEYVAVA 406
>BU550032 similar to GP|27901696|gb| gag-pol polyprotein {Vitis vinifera},
partial (21%)
Length = 698
Score = 94.7 bits (234), Expect = 9e-20
Identities = 41/81 (50%), Positives = 59/81 (72%)
Frame = -2
Query: 446 RTKKQLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAAN 505
++KKQ +V+ S++ EY ++ TCEL W+ LQ+L+ LYCDNQ+ALHIA+N
Sbjct: 697 KSKKQTVVA*XSAEVEY*SMTMVTCELMWIKQXLQELRFCEVVQMKLYCDNQAALHIASN 518
Query: 506 PIFHERTKHLEIDCHLVQDKL 526
P+FHER KH+EIDCH +++KL
Sbjct: 517 PVFHERAKHIEIDCHFIREKL 455
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.355 0.156 0.561
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,022,028
Number of Sequences: 63676
Number of extensions: 498162
Number of successful extensions: 6675
Number of sequences better than 10.0: 88
Number of HSP's better than 10.0 without gapping: 3768
Number of HSP's successfully gapped in prelim test: 344
Number of HSP's that attempted gapping in prelim test: 2796
Number of HSP's gapped (non-prelim): 4271
length of query: 527
length of database: 12,639,632
effective HSP length: 102
effective length of query: 425
effective length of database: 6,144,680
effective search space: 2611489000
effective search space used: 2611489000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC123574.1


