

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123571.8 - phase: 0
(522 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
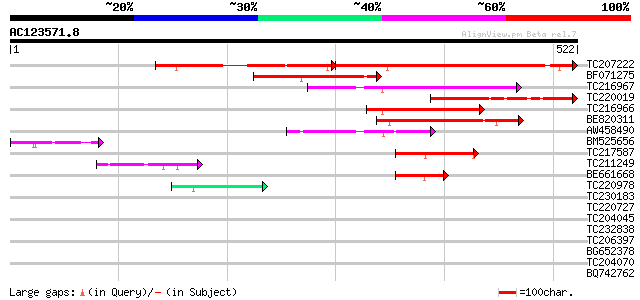
Score E
Sequences producing significant alignments: (bits) Value
TC207222 311 e-139
BF071275 similar to PIR|S14981|S149 extensin class I (clone w1-8... 164 9e-41
TC216967 158 6e-39
TC220019 131 6e-31
TC216966 weakly similar to UP|Q6PHL0 (Q6PHL0) Wu:fd15e07 protein... 126 3e-29
BE820311 123 2e-28
AW458490 weakly similar to GP|20147195|gb| AT5g66100/K2A18_18 {A... 107 1e-23
BM525656 82 8e-16
TC217587 similar to UP|Q94A38 (Q94A38) AT5g46250/MPL12_3, partia... 56 3e-08
TC211249 similar to UP|FXE3_MOUSE (Q9QY14) Forkhead box protein ... 54 1e-07
BE661668 similar to GP|10177706|dbj contains similarity to RNA-b... 46 4e-05
TC220978 similar to GB|AAP75802.1|32189295|BT009652 At2g35230 {A... 42 9e-04
TC230183 homologue to UP|GLHR_ANTEL (P35409) Probable glycoprote... 41 0.001
TC220727 40 0.003
TC204045 similar to UP|Q9M6R7 (Q9M6R7) Extensin, partial (23%) 40 0.003
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 40 0.003
TC206397 similar to GB|AAM91060.1|22136548|AY129474 At2g43970/F6... 39 0.004
BG652378 similar to PIR|T10863|T108 extensin precursor - kidney ... 39 0.004
TC204070 similar to UP|Q39835 (Q39835) Extensin, partial (34%) 39 0.004
BQ742762 PIR|T06782|T0 extensin - soybean, partial (32%) 39 0.006
>TC207222
Length = 2191
Score = 311 bits (798), Expect(2) = e-139
Identities = 159/227 (70%), Positives = 176/227 (77%), Gaps = 5/227 (2%)
Frame = +1
Query: 301 FIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPMP---LYYAGPDPQLH 357
F HP P+RP G +GFHELA P+V A PPPP DSLRGVPFVPPMP L++ GPDPQLH
Sbjct: 502 FFHPSPMRPIGSPIGFHELAPPLVFVAAPPPPPDSLRGVPFVPPMPHHPLFFTGPDPQLH 681
Query: 358 SKIVNQIDYYFSNENLVKDIFLRKNMDAQGWVPITLIAGFKKVMDLTDNIQLIIDAIRTS 417
+KIVNQ+DYYFSNENLVKD FLR+NMD QGWVPI LIAGF KVM LTDNIQ+I+DAI+TS
Sbjct: 682 NKIVNQVDYYFSNENLVKDTFLRQNMDDQGWVPIKLIAGFNKVMHLTDNIQVILDAIQTS 861
Query: 418 SVVEVQGDKIRRQNDWEKWIMPSPVQFPNVTSPEVLNQDMLAEKMRNIALETTIYDGAGG 477
SVVEVQGDKIRRQNDW +WIM PVQF N T+ VLN DMLAE++ NIALET+ YDGAGG
Sbjct: 862 SVVEVQGDKIRRQNDWRRWIMHPPVQFSNATTIGVLNPDMLAEQVHNIALETSDYDGAGG 1041
Query: 478 PVVLPDNSEHTPAFGDLSSPLQLSTSE--ITGEVDIHGSDFSTSRRN 522
P VLPD S H F D LQLSTSE G+V I GSD RN
Sbjct: 1042PDVLPDTSRHRSTFRD----LQLSTSEGPGPGQVGIQGSDHFIPARN 1170
Score = 204 bits (518), Expect(2) = e-139
Identities = 105/172 (61%), Positives = 119/172 (69%), Gaps = 5/172 (2%)
Frame = +3
Query: 135 GMESTPSSSMQRQVGDNV---NVNNMAPTRQKSIKHNSSNASSNGGHTQQSAPQVSIAAT 191
G+ ST SS QR+V DN N N++ P RQKS+KH+SSNASSNGGH Q S PQVSIAAT
Sbjct: 54 GLGSTSFSSSQREVSDNARTNNTNSLVPARQKSMKHHSSNASSNGGHAQHSVPQVSIAAT 233
Query: 192 GSHTSSSRDHTQSPRDHTQSPRDHAQSPRDHTQRSGFVPSDHPQQRNSFRHRNGGPHQRG 251
GS SS P+DHTQRSGF +DHPQQRNSFR+RNGG HQRG
Sbjct: 234 GSRNSS---------------------PKDHTQRSGFASNDHPQQRNSFRNRNGGQHQRG 350
Query: 252 DGSHHHHNYGNRRDQDWNSRRNYNGRDMHVPPRVSPRIIR--PSLPPNSAPF 301
DGS HHHNYGNRRDQ+WN+ ++ RD HVPPRV+PR IR P PPNSA F
Sbjct: 351 DGS-HHHNYGNRRDQEWNNNXSFGSRDTHVPPRVAPRFIRPPPPPPPNSAQF 503
>BF071275 similar to PIR|S14981|S149 extensin class I (clone w1-8 L) - tomato
(fragment), partial (10%)
Length = 389
Score = 164 bits (415), Expect = 9e-41
Identities = 79/125 (63%), Positives = 92/125 (73%), Gaps = 7/125 (5%)
Frame = +1
Query: 225 RSGFVPSDHPQQRNSFRHRNGG-PHQRGDGSHHHHNYGNRRDQD-----WNSRRNYNGRD 278
R+GF+P+DHP RNSFRHRNGG PHQRGDG HHH+NYG RRDQD WN+ RN+NGRD
Sbjct: 10 RAGFLPNDHPPHRNSFRHRNGGGPHQRGDGHHHHNNYGGRRDQDPGNQDWNNHRNFNGRD 189
Query: 279 MHVPPRVSPRIIRPSLPPNSAP-FIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLR 337
+ PR PR IRP PPN A F PPPLRP+GG +GF EL +V PPPP++S+R
Sbjct: 190 NFMSPRFGPRFIRPPPPPNPAQLFPPPPPLRPYGGSIGFTELPPQMVYV--PPPPLESMR 363
Query: 338 GVPFV 342
GVPFV
Sbjct: 364 GVPFV 378
>TC216967
Length = 1117
Score = 158 bits (399), Expect = 6e-39
Identities = 89/202 (44%), Positives = 120/202 (59%), Gaps = 5/202 (2%)
Frame = +1
Query: 275 NGRDMHV-PPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPI 333
N D HV R+ PR + PPN APF+ P P+RP GF E + P
Sbjct: 19 NTTDAHVNQQRMPPRGLMRPPPPNPAPFMGPQPMRPLANPAGFPEY------YYFPTLQF 180
Query: 334 DSLRGVPF---VPPMPLYYAGPDPQLHSKIVNQIDYYFSNENLVKDIFLRKNMDAQGWVP 390
+ G+PF PP ++Y + L + I NQIDYYFS+ NLVKD +LR NMD QGWVP
Sbjct: 181 EPFGGMPFFTHAPPPAMFYPVAETPLTNTIANQIDYYFSDANLVKDEYLRSNMDEQGWVP 360
Query: 391 ITLIAGFKKVMDLTDNIQLIIDAIRTSSVVEVQGDKIRRQNDWEKWIMPSPVQFPNVT-S 449
ITLIA F +V LT NI+LI+D++RTS+VVEVQGDK+RR N+W KW+ + ++ + + S
Sbjct: 361 ITLIASFPRVRSLTSNIKLILDSLRTSTVVEVQGDKLRRCNEWMKWLPSAQLRANSGSIS 540
Query: 450 PEVLNQDMLAEKMRNIALETTI 471
P + + LA I ++ I
Sbjct: 541 PSGSSSNNLAADFEKITVDEAI 606
>TC220019
Length = 582
Score = 131 bits (330), Expect = 6e-31
Identities = 76/135 (56%), Positives = 94/135 (69%)
Frame = +2
Query: 388 WVPITLIAGFKKVMDLTDNIQLIIDAIRTSSVVEVQGDKIRRQNDWEKWIMPSPVQFPNV 447
WV I LIAGFKKV LT+NIQ+++DA+RTSSVVEVQGDKIRR+NDW +WIMP+ Q PNV
Sbjct: 2 WVTINLIAGFKKVKYLTENIQIVLDAVRTSSVVEVQGDKIRRRNDWRRWIMPAG-QVPNV 178
Query: 448 TSPEVLNQDMLAEKMRNIALETTIYDGAGGPVVLPDNSEHTPAFGDLSSPLQLSTSEITG 507
+ + Q LAE+++NI LETT + AG +L D+ T FGD +S STSE T
Sbjct: 179 RGSQTVGQ--LAEQVQNITLETTNNNNAG---ILDDSQNRT--FGDSNSQYLNSTSEGTA 337
Query: 508 EVDIHGSDFSTSRRN 522
+V I SD S S RN
Sbjct: 338 QVGIQVSDHSISARN 382
>TC216966 weakly similar to UP|Q6PHL0 (Q6PHL0) Wu:fd15e07 protein (Fragment),
partial (10%)
Length = 1027
Score = 126 bits (316), Expect = 3e-29
Identities = 60/112 (53%), Positives = 79/112 (69%), Gaps = 3/112 (2%)
Frame = +1
Query: 329 PPPPIDSLRGVPF---VPPMPLYYAGPDPQLHSKIVNQIDYYFSNENLVKDIFLRKNMDA 385
P + G+PF PP ++Y + L + I NQIDYYFS+ NLVKD +LR NMD
Sbjct: 43 PTLQFEPFGGMPFFTHAPPPAMFYPVAETPLTNTIANQIDYYFSDANLVKDEYLRSNMDE 222
Query: 386 QGWVPITLIAGFKKVMDLTDNIQLIIDAIRTSSVVEVQGDKIRRQNDWEKWI 437
QGWVPI+LIA F +V LT NI+LI+D++RTS+ VEVQGDK+RR+ +W KW+
Sbjct: 223 QGWVPISLIASFPRVRSLTSNIKLILDSLRTSTFVEVQGDKLRRRTEWMKWL 378
>BE820311
Length = 680
Score = 123 bits (309), Expect = 2e-28
Identities = 69/141 (48%), Positives = 88/141 (61%), Gaps = 5/141 (3%)
Frame = -3
Query: 338 GVPFVPPMPLY---YAGPDPQLHSKIVNQIDYYFSNENLVKDIFLRKNMDAQGWVPITLI 394
G+PF P + L + IV QI+YYFS+ NLVKD FLR MD QGWVP+TLI
Sbjct: 672 GMPFFTHSPXXTPXXXAAESPLSNMIVYQIEYYFSDANLVKDAFLRSKMDEQGWVPVTLI 493
Query: 395 AGFKKVMDLTDNIQLIIDAIRTSSVVEVQGDKIRRQNDWEKWIMPSPVQFPN--VTSPEV 452
A F +V +LT NIQLI+D++RTS+VVEVQGDK+RR N+W +W +PS + N SP
Sbjct: 492 ADFPRVKNLTTNIQLILDSLRTSTVVEVQGDKLRRLNEWMRW-LPSAQRRSNSGSISPSG 316
Query: 453 LNQDMLAEKMRNIALETTIYD 473
+ L + I LE T D
Sbjct: 315 STHNNLTANFQTIILEETTAD 253
>AW458490 weakly similar to GP|20147195|gb| AT5g66100/K2A18_18 {Arabidopsis
thaliana}, partial (7%)
Length = 411
Score = 107 bits (268), Expect = 1e-23
Identities = 62/143 (43%), Positives = 82/143 (56%), Gaps = 6/143 (4%)
Frame = +3
Query: 256 HHHNYGNRRDQDWNSRRNY-NGRDMHVP-PRVSPRIIRPSLPPNSAPFIHPPPLRPFGGH 313
+H++YG+RRD R NY N RD +P PR+ PR + PPN+A F+ P P+ PF
Sbjct: 9 YHNSYGSRRDHQ--DRGNYVNARDAFLPQPRMPPRGLLRHPPPNTAAFVGPQPIGPFANP 182
Query: 314 MGFHELAAPVVLFAGPPPPIDSLRGVPFV----PPMPLYYAGPDPQLHSKIVNQIDYYFS 369
+ F E + P ++ G+PF PP P + A P L + IV QI+YYFS
Sbjct: 183 ISFPEF------YYYQPVTVEQFTGMPFFTHSPPPTPFFSAAESP-LSNMIVYQIEYYFS 341
Query: 370 NENLVKDIFLRKNMDAQGWVPIT 392
+ NLVKD FLR MD QGWVP+T
Sbjct: 342 DANLVKDAFLRSKMDEQGWVPVT 410
>BM525656
Length = 430
Score = 81.6 bits (200), Expect = 8e-16
Identities = 52/93 (55%), Positives = 56/93 (59%), Gaps = 7/93 (7%)
Frame = +2
Query: 1 MEMIGNHSSNHSPDNLQSRR---LP---PWNQVVRGESESIAAVPAVSLSEESFPIVTAP 54
M M GNHS HS DN QSRR LP PWNQVV GESE +AAVP S S E FP AP
Sbjct: 182 MAMTGNHSPRHSSDNHQSRRATRLPASSPWNQVVCGESEPVAAVP--SSSTEDFPSAAAP 355
Query: 55 VDD-STSAEVISDNADNGGERNGGTGKRPAWNR 86
V+D S+SA SD NGG KRP WN+
Sbjct: 356 VEDFSSSATESSD--------NGGAAKRPVWNK 430
>TC217587 similar to UP|Q94A38 (Q94A38) AT5g46250/MPL12_3, partial (27%)
Length = 724
Score = 56.2 bits (134), Expect = 3e-08
Identities = 31/80 (38%), Positives = 51/80 (63%), Gaps = 4/80 (5%)
Frame = +2
Query: 356 LHSKIVNQIDYYFSNENLVKDIFLRK--NMDAQGWVPITLIAGFKKVMDLTDNIQLIIDA 413
L KI+ Q +YYFS+ENL D +L + +G+VP+++IA F+K+ LT + I A
Sbjct: 350 LKLKIIKQAEYYFSDENLPTDKYLLGFVKRNKEGFVPVSVIASFRKIKKLTRDHAFIAAA 529
Query: 414 IRTSSVVEVQGD--KIRRQN 431
++ SS++ V GD +++R N
Sbjct: 530 LKESSLLVVSGDGRRVKRLN 589
>TC211249 similar to UP|FXE3_MOUSE (Q9QY14) Forkhead box protein E3, partial
(5%)
Length = 536
Score = 54.3 bits (129), Expect = 1e-07
Identities = 37/108 (34%), Positives = 53/108 (48%), Gaps = 11/108 (10%)
Frame = +1
Query: 81 RPAWNRSSGNGGVSEVQPVMDAHSWPALSDSARGSTKSESSKGLLDGSSVSPWQGMESTP 140
+PAW + NG VSEV PVM AHSWP LS+S + + K S + + + + + +TP
Sbjct: 4 KPAWKKPESNG-VSEVGPVMGAHSWPDLSESTKPTAKLTSDSSV---KTAADEESLTTTP 171
Query: 141 -----SSSMQRQVGDNVN------VNNMAPTRQKSIKHNSSNASSNGG 177
S S Q+Q N N +N RQ+S+K + N G
Sbjct: 172 QDPVNSDSPQKQAIGNANPSPTPAMNYGMANRQRSMKQRGGSNGYNVG 315
>BE661668 similar to GP|10177706|dbj contains similarity to RNA-binding
protein~gene_id:MPL12.3 {Arabidopsis thaliana}, partial
(13%)
Length = 554
Score = 46.2 bits (108), Expect = 4e-05
Identities = 22/51 (43%), Positives = 35/51 (68%), Gaps = 2/51 (3%)
Frame = +1
Query: 356 LHSKIVNQIDYYFSNENLVKDIFLR--KNMDAQGWVPITLIAGFKKVMDLT 404
L KI+ Q++YYFS+ENL D +L + +G+VP+++IA F+K+ LT
Sbjct: 343 LKLKIIKQVEYYFSDENLPTDKYLLGFVKRNKEGFVPVSVIASFRKIKKLT 495
Score = 28.9 bits (63), Expect = 5.8
Identities = 17/60 (28%), Positives = 28/60 (46%), Gaps = 5/60 (8%)
Frame = -2
Query: 234 PQQRNSFRHRNGGPHQRGDGSHHHHNYGNRRDQDWNSRRN-----YNGRDMHVPPRVSPR 288
P++R+ R G H R H H + + RD+D + R+ + R+ VP + PR
Sbjct: 322 PRRRDHRGPRGGHAHARDRARGHGHAHAHARDRDRDRDRDRALRLKSAREFQVPQQRRPR 143
>TC220978 similar to GB|AAP75802.1|32189295|BT009652 At2g35230 {Arabidopsis
thaliana;} , partial (15%)
Length = 732
Score = 41.6 bits (96), Expect = 9e-04
Identities = 28/93 (30%), Positives = 37/93 (39%), Gaps = 5/93 (5%)
Frame = +2
Query: 150 DNVNVNNMAPTRQKSIKH-----NSSNASSNGGHTQQSAPQVSIAATGSHTSSSRDHTQS 204
DN+ VN M +KS H N++N+S NGG QQ PQ + RD Q
Sbjct: 179 DNLGVNKMGKNIRKSPLHQPNYGNNNNSSMNGGRQQQQQPQPQPQVYNISKNDFRDIVQQ 358
Query: 205 PRDHTQSPRDHAQSPRDHTQRSGFVPSDHPQQR 237
+DH QS Q + P Q+
Sbjct: 359 LTGSPSQSQDHPQSKPPQNQNNSPKPQSMRLQK 457
>TC230183 homologue to UP|GLHR_ANTEL (P35409) Probable glycoprotein hormone
G-protein coupled receptor precursor, partial (6%)
Length = 878
Score = 40.8 bits (94), Expect = 0.001
Identities = 28/95 (29%), Positives = 40/95 (41%), Gaps = 2/95 (2%)
Frame = -2
Query: 167 HNSSNASSNGGHTQQSAPQVSIAATGSHTSSSRDHTQSPRDHTQSPRDHAQSPRDHTQRS 226
H + S H+Q S S + H S QS +S R +Q H Q
Sbjct: 439 HRHQSQGSRRHHSQGSRRHQSQGSRRHHQSQGSRRHQSQ----ESRRHQSQGTCRHHQSQ 272
Query: 227 GFVPSDHPQQRNSFRHRNGGPHQRGDGSHH--HHN 259
G H Q + S++H++ G H + GSHH HH+
Sbjct: 271 G--SRRHHQSQGSYQHQSQGSHHQSQGSHHQSHHS 173
Score = 31.6 bits (70), Expect = 0.90
Identities = 17/45 (37%), Positives = 22/45 (48%), Gaps = 2/45 (4%)
Frame = -2
Query: 231 SDHPQQRNSFRHRNGGP--HQRGDGSHHHHNYGNRRDQDWNSRRN 273
S Q + S RH + G HQ HH + G+RR Q SRR+
Sbjct: 442 SHRHQSQGSRRHHSQGSRRHQSQGSRRHHQSQGSRRHQSQESRRH 308
Score = 29.3 bits (64), Expect = 4.5
Identities = 11/28 (39%), Positives = 17/28 (60%)
Frame = -2
Query: 250 RGDGSHHHHNYGNRRDQDWNSRRNYNGR 277
+ GS HH+ G+RR Q SRR++ +
Sbjct: 430 QSQGSRRHHSQGSRRHQSQGSRRHHQSQ 347
>TC220727
Length = 479
Score = 40.0 bits (92), Expect = 0.003
Identities = 25/99 (25%), Positives = 50/99 (50%), Gaps = 16/99 (16%)
Frame = +1
Query: 353 DPQLHSKIVNQIDYYFSNENLVKDIFLRKNM--DAQGWVPITLIAGF---KKVMDLTD-- 405
D + K++ Q+++YF + NL+ D F+RK++ G + + LI F +K ++L D
Sbjct: 76 DEETTKKVIRQVEFYFGDSNLLTDGFMRKSITESEDGMISLALICSFNRMRKHLNLGDVK 255
Query: 406 -------NIQLIIDAIRTSSVVEV--QGDKIRRQNDWEK 435
+ + +R S+ ++V G K+ R+ + K
Sbjct: 256 PDEVAQETVNTVAQTLRNSASLKVSEDGKKVGRKTELPK 372
>TC204045 similar to UP|Q9M6R7 (Q9M6R7) Extensin, partial (23%)
Length = 650
Score = 39.7 bits (91), Expect = 0.003
Identities = 24/82 (29%), Positives = 35/82 (42%), Gaps = 1/82 (1%)
Frame = +2
Query: 283 PRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFV 342
P P + + PP P PPP + + PV + PPPP+ S
Sbjct: 35 PSPPPPVYKYKSPPYKYPSPPPPPYK-------YPSPPPPVYKYKSPPPPVHS------- 172
Query: 343 PPMPLY-YAGPDPQLHSKIVNQ 363
PP P+Y YA P P H ++++
Sbjct: 173 PPPPVYIYASPPPPYHY*VISE 238
Score = 29.6 bits (65), Expect = 3.4
Identities = 21/59 (35%), Positives = 26/59 (43%), Gaps = 8/59 (13%)
Frame = +2
Query: 282 PPRV----SPRIIRPSLPPNSAPFIHPPPLRPFGGHMG----FHELAAPVVLFAGPPPP 332
PP V SP PS PP P+ +P P P + H PV ++A PPPP
Sbjct: 44 PPPVYKYKSPPYKYPSPPP--PPYKYPSPPPPVYKYKSPPPPVHSPPPPVYIYASPPPP 214
Score = 28.1 bits (61), Expect = 9.9
Identities = 12/40 (30%), Positives = 18/40 (45%), Gaps = 4/40 (10%)
Frame = +2
Query: 322 PVVLFAGPPPPIDSLRGVPF----VPPMPLYYAGPDPQLH 357
P + PPPP+ + P+ PP P Y P P ++
Sbjct: 20 PPYKYPSPPPPVYKYKSPPYKYPSPPPPPYKYPSPPPPVY 139
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 39.7 bits (91), Expect = 0.003
Identities = 44/178 (24%), Positives = 77/178 (42%), Gaps = 3/178 (1%)
Frame = -3
Query: 24 WNQVVRGESESIAAVPAVSLSEESFPIVTAPVDDSTSAEVISDNADNGGERNGGTGKRPA 83
W Q+ R E S ++ P+ S + + S S ++ + + G+ A
Sbjct: 798 WTQLYRLEGLSSSSPPSSSSESSLSDLFLFGGASAFSQSSSSTSSSSSSPFSSGSKSLDA 619
Query: 84 WNRSS-GNGGVSEVQPVMDAHSWPALSDSARGSTKSESSKGLLDGSSVSP--WQGMESTP 140
+ S+ G + P + + S S+ S+ SESS L SS SP W S+
Sbjct: 618 SSSSAVSKGRIRYSVPKSSSSPCSSSSSSSSSSSSSESSSESLSSSSGSPPRWTSPSSSS 439
Query: 141 SSSMQRQVGDNVNVNNMAPTRQKSIKHNSSNASSNGGHTQQSAPQVSIAATGSHTSSS 198
SSS + + + A + S+ SS++SS+ + SA + ++T S++SSS
Sbjct: 438 SSS------SSSSPPSGASSSSSSLSSPSSSSSSSSSSSSSSASSTT-SSTDSYSSSS 286
>TC206397 similar to GB|AAM91060.1|22136548|AY129474 At2g43970/F6E13.10
{Arabidopsis thaliana;} , partial (41%)
Length = 1881
Score = 39.3 bits (90), Expect = 0.004
Identities = 21/51 (41%), Positives = 31/51 (60%), Gaps = 2/51 (3%)
Frame = +1
Query: 382 NMDAQGWVPITLIAGFKKVMDLTDNIQLIIDAIRTSS--VVEVQGDKIRRQ 430
N D +G+VPI+++A FKK+ L + + +R SS VV G KI+RQ
Sbjct: 751 NKDPEGFVPISVVASFKKIKALIASHSQLATVLRNSSKLVVSEDGKKIKRQ 903
>BG652378 similar to PIR|T10863|T108 extensin precursor - kidney bean,
partial (14%)
Length = 361
Score = 39.3 bits (90), Expect = 0.004
Identities = 22/76 (28%), Positives = 28/76 (35%)
Frame = +3
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF 341
PP P PP P+ +P P P + H P ++ PPPP
Sbjct: 123 PPPKKPYYYHSPPPPPKKPYKYPSPPPPVHPYPHPHPHPHPPYVYHSPPPP--------- 275
Query: 342 VPPMPLYYAGPDPQLH 357
P P YY P P +H
Sbjct: 276 -PKKPYYYHSPPPPVH 320
Score = 30.8 bits (68), Expect = 1.5
Identities = 22/74 (29%), Positives = 25/74 (33%), Gaps = 4/74 (5%)
Frame = +3
Query: 292 PSLPPNSAPFIHPPPLRPFGGHMGFHELAAPV----VLFAGPPPPIDSLRGVPFVPPMPL 347
PS PP + H PP P +H P + PPPP P PP
Sbjct: 33 PSPPPPMPYYYHSPPPPPKKKPYYYHSPPPPPPKKPYYYHSPPPPPKKPYKYPSPPPPVH 212
Query: 348 YYAGPDPQLHSKIV 361
Y P P H V
Sbjct: 213 PYPHPHPHPHPPYV 254
>TC204070 similar to UP|Q39835 (Q39835) Extensin, partial (34%)
Length = 596
Score = 39.3 bits (90), Expect = 0.004
Identities = 27/94 (28%), Positives = 35/94 (36%)
Frame = +2
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF 341
PP + P PP P+ +P P P P + PPPP P
Sbjct: 317 PPPPVYKYKSPPPPPPPPPYKYPSPPPP------------PAYYYKSPPPPPKKPYKYPS 460
Query: 342 VPPMPLYYAGPDPQLHSKIVNQIDYYFSNENLVK 375
PP YA P P H+ IV + + +NL K
Sbjct: 461 PPPPHYVYASPPPPYHN*IVILV*HPMDLKNLCK 562
Score = 35.8 bits (81), Expect = 0.048
Identities = 22/73 (30%), Positives = 29/73 (39%), Gaps = 1/73 (1%)
Frame = +2
Query: 283 PRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFV 342
P P + + PP + PPP P + PV + PPPP+ + P
Sbjct: 185 PSPPPPVYKYKSPPPPYKYSSPPP--PPKKPYKYPSPPPPVYKYKSPPPPVYKYKSPPPP 358
Query: 343 PPMPLY-YAGPDP 354
PP P Y Y P P
Sbjct: 359 PPPPPYKYPSPPP 397
Score = 32.7 bits (73), Expect = 0.40
Identities = 20/76 (26%), Positives = 29/76 (37%)
Frame = +2
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF 341
PP P PP P+ +P P P + ++ P ++ PPPP P
Sbjct: 119 PPEKPYNYHSPPPPPPKKPYKYPSPPPP----VYKYKSPPPPYKYSSPPPPPKKPYKYPS 286
Query: 342 VPPMPLYYAGPDPQLH 357
PP Y P P ++
Sbjct: 287 PPPPVYKYKSPPPPVY 334
>BQ742762 PIR|T06782|T0 extensin - soybean, partial (32%)
Length = 420
Score = 38.9 bits (89), Expect = 0.006
Identities = 27/80 (33%), Positives = 34/80 (41%), Gaps = 4/80 (5%)
Frame = +3
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVP- 340
PP+ SP PS PP + PPP P+ + P + PPPP+ + P
Sbjct: 198 PPKHSPPYKYPSPPPPVYKYKSPPP--PY----KYPSPPPPPYKYPSPPPPVYKYKSPPP 359
Query: 341 --FVPPMPLY-YAGPDPQLH 357
PP P Y YA P P H
Sbjct: 360 PVHSPPPPHYIYASPPPPYH 419
Score = 30.8 bits (68), Expect = 1.5
Identities = 14/34 (41%), Positives = 17/34 (49%)
Frame = +3
Query: 325 LFAGPPPPIDSLRGVPFVPPMPLYYAGPDPQLHS 358
+++ PPPP S PP P YY P P HS
Sbjct: 84 IYSSPPPPKHS-------PPPPYYYHSPPPPKHS 164
Score = 30.0 bits (66), Expect = 2.6
Identities = 15/37 (40%), Positives = 16/37 (42%)
Frame = +3
Query: 322 PVVLFAGPPPPIDSLRGVPFVPPMPLYYAGPDPQLHS 358
P + PPPP S PP P YY P P HS
Sbjct: 123 PPYYYHSPPPPKHS-------PPPPYYYHSPPPPKHS 212
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.130 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,022,989
Number of Sequences: 63676
Number of extensions: 463036
Number of successful extensions: 3990
Number of sequences better than 10.0: 291
Number of HSP's better than 10.0 without gapping: 3234
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3697
length of query: 522
length of database: 12,639,632
effective HSP length: 102
effective length of query: 420
effective length of database: 6,144,680
effective search space: 2580765600
effective search space used: 2580765600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC123571.8


