

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122727.11 + phase: 0 /pseudo/partial
(359 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
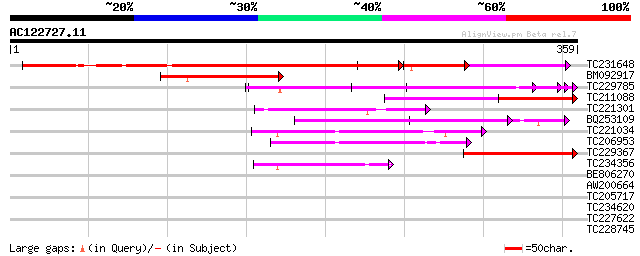
Score E
Sequences producing significant alignments: (bits) Value
TC231648 weakly similar to UP|Q6K7E2 (Q6K7E2) Mitochondrial tran... 323 e-106
BM092917 similar to GP|3212859|gb|A expressed protein {Arabidops... 87 2e-17
TC229785 weakly similar to UP|Q6K5F5 (Q6K5F5) Mitochondrial tran... 85 6e-17
TC211088 similar to UP|Q6K7E2 (Q6K7E2) Mitochondrial transcripti... 56 3e-08
TC221301 weakly similar to UP|Q9SJ50 (Q9SJ50) At2g36000/F11F19.9... 55 6e-08
BQ253109 54 1e-07
TC221034 weakly similar to UP|O80618 (O80618) Predicted by genef... 51 9e-07
TC206953 similar to UP|O80618 (O80618) Predicted by genefinder a... 49 4e-06
TC229367 similar to GB|AAP75804.1|32189299|BT009654 At4g09620 {A... 44 1e-04
TC234356 weakly similar to GB|AAN18117.1|23308295|BT000548 At4g3... 44 1e-04
BE806270 31 0.99
AW200664 similar to GP|9971154|dbj mkpA {Dictyostelium discoideu... 30 2.2
TC205717 homologue to UP|O65463 (O65463) Protein kinase ADK1-lik... 30 2.2
TC234620 weakly similar to GB|AAO63444.1|28951041|BT005380 At1g6... 29 3.8
TC227622 similar to UP|Q7XJT9 (Q7XJT9) NAM (No apical meristem)-... 28 4.9
TC228745 similar to UP|O65201 (O65201) Acyl-CoA oxidase , parti... 28 8.4
>TC231648 weakly similar to UP|Q6K7E2 (Q6K7E2) Mitochondrial transcription
termination factor-like, partial (12%)
Length = 903
Score = 323 bits (827), Expect(2) = e-106
Identities = 174/241 (72%), Positives = 198/241 (81%)
Frame = +1
Query: 9 LKNMNFSGTITKPNFPFPPTEMPILTFGHSKLTAATLALQLPCNYIRKIRFITKLQCSVA 68
+K ++FSG ITKP F TEMPILTF H KL+A TLAL R++ IT+LQ SVA
Sbjct: 88 MKVISFSGGITKPCFFIAQTEMPILTFAHCKLSA-TLAL-------RRMGSITRLQYSVA 243
Query: 69 GRTDSYRTSAADSQGSKRDSGEIQRKRRGASSLYAGYARPSLSEMKKDKATLRKVVYEFL 128
RT + + + D SK DSG + R+R G SS + Y+RPSLSEMKK+KA +R+ VYEFL
Sbjct: 244 DRT--FTSGSVDLPASKTDSGRVNRRRGGGSS--SVYSRPSLSEMKKEKAAIREKVYEFL 411
Query: 129 RGIGIVPDELDGLELPVTVDVMKERVDFLHSLGLTIEDINNYPLVLGCSVKKNMVPVLDY 188
R IGIVPDELDGLELPVTVDVM+ERVDFLHSLGLTIEDINNYPLVLGCSVKKNM+PVLDY
Sbjct: 412 RAIGIVPDELDGLELPVTVDVMRERVDFLHSLGLTIEDINNYPLVLGCSVKKNMIPVLDY 591
Query: 189 LGKLGVRKSTITQFLRTYPQVLHASVVVDLVPVVKYLQGMDIKPDDIPRVLERYPEVLGF 248
LGKLGVRKS+ITQFL+ YPQVLHASVVVDL+PVV YL+GMDIK DD+PRVLERYPEVLGF
Sbjct: 592 LGKLGVRKSSITQFLQRYPQVLHASVVVDLMPVVNYLKGMDIKFDDVPRVLERYPEVLGF 771
Query: 249 K 249
K
Sbjct: 772 K 774
Score = 81.6 bits (200), Expect(2) = e-106
Identities = 39/42 (92%), Positives = 42/42 (99%)
Frame = +2
Query: 250 LEGTMSTSVAYLIGIGVGRRELGGILTRFPEILGMRVGRVIK 291
LEGTMSTSVAYLIGIGVGRRE+GG+LTR+PEILGMRVGRVIK
Sbjct: 776 LEGTMSTSVAYLIGIGVGRREIGGVLTRYPEILGMRVGRVIK 901
Score = 46.2 bits (108), Expect = 2e-05
Identities = 30/138 (21%), Positives = 68/138 (48%), Gaps = 3/138 (2%)
Frame = +1
Query: 221 VVKYLQGMDIKPDDIPRVLERYPEVLGFKLEGT---MSTSVAYLIGIGVGRRELGGILTR 277
V ++L+ + I PD++ G +L T M V +L +G+ ++
Sbjct: 397 VYEFLRAIGIVPDELD----------GLELPVTVDVMRERVDFLHSLGLTIEDINN---- 534
Query: 278 FPEILGMRVGRVIKPFVEYLESLGIPRLAIARLIETQPYILGFDLDEKVKPNVKSLEEFN 337
+P +LG V + + P ++YL LG+ + +I + ++ P +L + + P V L+ +
Sbjct: 535 YPLVLGCSVKKNMIPVLDYLGKLGVRKSSITQFLQRYPQVLHASVVVDLMPVVNYLKGMD 714
Query: 338 VRETSLASIIAQYPDIIG 355
++ + ++ +YP+++G
Sbjct: 715 IKFDDVPRVLERYPEVLG 768
Score = 28.9 bits (63), Expect = 3.8
Identities = 13/29 (44%), Positives = 19/29 (64%)
Frame = +2
Query: 222 VKYLQGMDIKPDDIPRVLERYPEVLGFKL 250
V YL G+ + +I VL RYPE+LG ++
Sbjct: 800 VAYLIGIGVGRREIGGVLTRYPEILGMRV 886
>BM092917 similar to GP|3212859|gb|A expressed protein {Arabidopsis
thaliana}, partial (15%)
Length = 421
Score = 86.7 bits (213), Expect = 2e-17
Identities = 40/82 (48%), Positives = 61/82 (73%), Gaps = 4/82 (4%)
Frame = +2
Query: 96 RGASSLYAGYARPSLS----EMKKDKATLRKVVYEFLRGIGIVPDELDGLELPVTVDVMK 151
+ ++S Y PS++ + +K+K R +++++L+G+GI+PDEL LELP TVDVM+
Sbjct: 176 QSSASKLPEYEMPSVTWGVIQGRKEKLVSRVIIFDYLKGLGIIPDELHDLELPSTVDVMR 355
Query: 152 ERVDFLHSLGLTIEDINNYPLV 173
ERV+FL LGLT++DINNYPL+
Sbjct: 356 ERVEFLQKLGLTVDDINNYPLM 421
>TC229785 weakly similar to UP|Q6K5F5 (Q6K5F5) Mitochondrial transcription
termination factor-like protein, partial (74%)
Length = 1351
Score = 84.7 bits (208), Expect = 6e-17
Identities = 52/205 (25%), Positives = 102/205 (49%), Gaps = 4/205 (1%)
Frame = +3
Query: 150 MKERVDFLHSLGLTIEDINN----YPLVLGCSVKKNMVPVLDYLGKLGVRKSTITQFLRT 205
++ + +L LG+ IE I + +P S++ + PV+++ +LGV K I L
Sbjct: 114 LRPHIVYLMELGMDIEQIRSITRRFPSFAYYSLEGKIKPVVEFFLELGVPKENIPTILTK 293
Query: 206 YPQVLHASVVVDLVPVVKYLQGMDIKPDDIPRVLERYPEVLGFKLEGTMSTSVAYLIGIG 265
PQ+ S+ +L P +K+ + + + + P+V+ R+P +L + M S+ +L+ +G
Sbjct: 294 RPQLCGISLSENLKPTMKFFESLGVDKNQWPKVIYRFPALLTYSRPKVME-SIDFLLELG 470
Query: 266 VGRRELGGILTRFPEILGMRVGRVIKPFVEYLESLGIPRLAIARLIETQPYILGFDLDEK 325
+ +G ILTR P I+ V ++P +Y SLG+ + L+ P G ++
Sbjct: 471 LSEESIGKILTRCPNIVSYSVEDNLRPTAKYFHSLGV---EVGVLLFRCPQNFGLSIENN 641
Query: 326 VKPNVKSLEEFNVRETSLASIIAQY 350
+KP + E + ++I++Y
Sbjct: 642 LKPATEFFLERGYTLEEIGTMISRY 716
Score = 67.8 bits (164), Expect = 7e-12
Identities = 37/143 (25%), Positives = 75/143 (51%)
Frame = +3
Query: 217 DLVPVVKYLQGMDIKPDDIPRVLERYPEVLGFKLEGTMSTSVAYLIGIGVGRRELGGILT 276
+L P + YL + + + I + R+P + LEG + V + + +GV + + ILT
Sbjct: 111 NLRPHIVYLMELGMDIEQIRSITRRFPSFAYYSLEGKIKPVVEFFLELGVPKENIPTILT 290
Query: 277 RFPEILGMRVGRVIKPFVEYLESLGIPRLAIARLIETQPYILGFDLDEKVKPNVKSLEEF 336
+ P++ G+ + +KP +++ ESLG+ + ++I P +L + KV ++ L E
Sbjct: 291 KRPQLCGISLSENLKPTMKFFESLGVDKNQWPKVIYRFPALLTYS-RPKVMESIDFLLEL 467
Query: 337 NVRETSLASIIAQYPDIIGTDLE 359
+ E S+ I+ + P+I+ +E
Sbjct: 468 GLSEESIGKILTRCPNIVSYSVE 536
Score = 59.3 bits (142), Expect = 3e-09
Identities = 28/103 (27%), Positives = 51/103 (49%)
Frame = +3
Query: 252 GTMSTSVAYLIGIGVGRRELGGILTRFPEILGMRVGRVIKPFVEYLESLGIPRLAIARLI 311
G + + YL+ +G+ ++ I RFP + IKP VE+ LG+P+ I ++
Sbjct: 108 GNLRPHIVYLMELGMDIEQIRSITRRFPSFAYYSLEGKIKPVVEFFLELGVPKENIPTIL 287
Query: 312 ETQPYILGFDLDEKVKPNVKSLEEFNVRETSLASIIAQYPDII 354
+P + G L E +KP +K E V + +I ++P ++
Sbjct: 288 TKRPQLCGISLSENLKPTMKFFESLGVDKNQWPKVIYRFPALL 416
Score = 53.1 bits (126), Expect = 2e-07
Identities = 45/187 (24%), Positives = 81/187 (43%), Gaps = 4/187 (2%)
Frame = +3
Query: 152 ERVDFLHSLGLTIEDINNY----PLVLGCSVKKNMVPVLDYLGKLGVRKSTITQFLRTYP 207
E +DFL LGL+ E I P ++ SV+ N+ P Y LGV + L P
Sbjct: 441 ESIDFLLELGLSEESIGKILTRCPNIVSYSVEDNLRPTAKYFHSLGVEVGVL---LFRCP 611
Query: 208 QVLHASVVVDLVPVVKYLQGMDIKPDDIPRVLERYPEVLGFKLEGTMSTSVAYLIGIGVG 267
Q S+ +L P ++ ++I ++ RY + F L + + + G
Sbjct: 612 QNFGLSIENNLKPATEFFLERGYTLEEIGTMISRYGALYTFSLTENLIPKWDFFLTTGYP 791
Query: 268 RRELGGILTRFPEILGMRVGRVIKPFVEYLESLGIPRLAIARLIETQPYILGFDLDEKVK 327
+ EL +FP+ G + IKP E + G+ +L + +++ + + DE +K
Sbjct: 792 KSEL----VKFPQYFGYNLEERIKPRFEIMTKSGV-KLLLNQVLS----LSSSNFDEALK 944
Query: 328 PNVKSLE 334
+K ++
Sbjct: 945 KKMKKMQ 965
>TC211088 similar to UP|Q6K7E2 (Q6K7E2) Mitochondrial transcription
termination factor-like, partial (38%)
Length = 862
Score = 55.8 bits (133), Expect = 3e-08
Identities = 26/50 (52%), Positives = 35/50 (70%)
Frame = +2
Query: 310 LIETQPYILGFDLDEKVKPNVKSLEEFNVRETSLASIIAQYPDIIGTDLE 359
++E + Y+ G+DL+E VKPNV+ L F V LASIIAQYP I+G L+
Sbjct: 2 MLEKRAYVXGYDLEETVKPNVECLISFGVGRDCLASIIAQYPQILGLPLK 151
Score = 49.7 bits (117), Expect = 2e-06
Identities = 33/123 (26%), Positives = 61/123 (48%), Gaps = 1/123 (0%)
Frame = +2
Query: 238 VLERYPEVLGFKLEGTMSTSVAYLIGIGVGRRELGGILTRFPEILGMRVGRVIKPFVEYL 297
+LE+ V G+ LE T+ +V LI GVGR L I+ ++P+ILG+ + + +
Sbjct: 2 MLEKRAYVXGYDLEETVKPNVECLISFGVGRDCLASIIAQYPQILGLPLKAKLSTQQYFF 181
Query: 298 E-SLGIPRLAIARLIETQPYILGFDLDEKVKPNVKSLEEFNVRETSLASIIAQYPDIIGT 356
L + AR++E P ++ +KP V+ L + +AS++ + P ++
Sbjct: 182 SLKLKVDPEGFARVVENMPQVVSLHQHVIMKP-VEFLLGRTIPAQDVASMVVKCPQLVAL 358
Query: 357 DLE 359
+E
Sbjct: 359 RVE 367
Score = 38.9 bits (89), Expect = 0.004
Identities = 39/160 (24%), Positives = 70/160 (43%), Gaps = 5/160 (3%)
Frame = +2
Query: 148 DVMKERVDFLHSLGL----TIEDINNYPLVLGCSVKKNMVPVLDYLG-KLGVRKSTITQF 202
+ +K V+ L S G+ I YP +LG +K + + KL V +
Sbjct: 44 ETVKPNVECLISFGVGRDCLASIIAQYPQILGLPLKAKLSTQQYFFSLKLKVDPEGFARV 223
Query: 203 LRTYPQVLHASVVVDLVPVVKYLQGMDIKPDDIPRVLERYPEVLGFKLEGTMSTSVAYLI 262
+ PQV+ V + PV ++L G I D+ ++ + P+++ ++E ++ +
Sbjct: 224 VENMPQVVSLHQHVIMKPV-EFLLGRTIPAQDVASMVVKCPQLVALRVELMKNSYYFFKS 400
Query: 263 GIGVGRRELGGILTRFPEILGMRVGRVIKPFVEYLESLGI 302
+G +EL FPE + IKP + L+S GI
Sbjct: 401 EMGRPLQEL----VEFPEYFTYSLESRIKPRYQRLKSKGI 508
Score = 37.7 bits (86), Expect = 0.008
Identities = 22/105 (20%), Positives = 51/105 (47%), Gaps = 1/105 (0%)
Frame = +2
Query: 220 PVVKYLQGMDIKPDDIPRVLERYPEVLGFKLEGTMSTSVAYL-IGIGVGRRELGGILTRF 278
P V+ L + D + ++ +YP++LG L+ +ST + + + V ++
Sbjct: 56 PNVECLISFGVGRDCLASIIAQYPQILGLPLKAKLSTQQYFFSLKLKVDPEGFARVVENM 235
Query: 279 PEILGMRVGRVIKPFVEYLESLGIPRLAIARLIETQPYILGFDLD 323
P+++ + ++KP VE+L IP +A ++ P ++ ++
Sbjct: 236 PQVVSLHQHVIMKP-VEFLLGRTIPAQDVASMVVKCPQLVALRVE 367
>TC221301 weakly similar to UP|Q9SJ50 (Q9SJ50) At2g36000/F11F19.9, partial
(48%)
Length = 835
Score = 54.7 bits (130), Expect = 6e-08
Identities = 33/114 (28%), Positives = 55/114 (47%), Gaps = 3/114 (2%)
Frame = +2
Query: 156 FLHSLGLTIEDINNYPLVLGCSVKKNMVPVLDYLGKLGVRKSTITQFLRTYPQVLHASVV 215
FL S+G IE++N + +L CSV++ +P +DY +G + T R +PQ+ S+
Sbjct: 110 FLQSIG--IEEVNKHTDLLSCSVEEKFMPRIDYFENIGFSRRDATSMFRRFPQLFCYSIK 283
Query: 216 VDLVPVVKYL---QGMDIKPDDIPRVLERYPEVLGFKLEGTMSTSVAYLIGIGV 266
+L P Y G D+K L+ +P+ F LE + + +GV
Sbjct: 284 NNLEPKYSYFVVEMGRDLKE------LKEFPQYFSFSLENRIKPRHKQCVEMGV 427
>BQ253109
Length = 421
Score = 53.9 bits (128), Expect = 1e-07
Identities = 34/140 (24%), Positives = 73/140 (51%), Gaps = 2/140 (1%)
Frame = +1
Query: 181 NMVPVLDYLGKLGVRKSTITQFLRTYPQVLHASVVVDLVPVVKYL-QGMDIKPDDIPRVL 239
N+ + +L LG+ S I Q + P + SV L P V+YL + + IK D+ +V+
Sbjct: 1 NLKSRVAFLRGLGIPNSRIGQIIAAAPSLFSYSVENSLKPTVRYLIEEVGIKEKDLGKVI 180
Query: 240 ERYPEVLGFKLEGTMSTSVAYLI-GIGVGRRELGGILTRFPEILGMRVGRVIKPFVEYLE 298
+ P++L +++ + +T +L +G R + ++T+ P++L + + P + +L
Sbjct: 181 QLSPQILVQRIDISWNTRCMFLTKELGAPRDSIVKMVTKHPQLLHYSIDDGLLPRINFLR 360
Query: 299 SLGIPRLAIARLIETQPYIL 318
S+G+ I +++ + +L
Sbjct: 361 SIGMKNSDIVKVLTSLTQVL 420
Score = 43.9 bits (102), Expect = 1e-04
Identities = 26/105 (24%), Positives = 52/105 (48%), Gaps = 4/105 (3%)
Frame = +1
Query: 254 MSTSVAYLIGIGVGRRELGGILTRFPEILGMRVGRVIKPFVEYL-ESLGIPRLAIARLIE 312
+ + VA+L G+G+ +G I+ P + V +KP V YL E +GI + ++I+
Sbjct: 4 LKSRVAFLRGLGIPNSRIGQIIAAAPSLFSYSVENSLKPTVRYLIEEVGIKEKDLGKVIQ 183
Query: 313 TQPYILGFDLDEKVKPNVKSL---EEFNVRETSLASIIAQYPDII 354
P IL +D + N + + +E S+ ++ ++P ++
Sbjct: 184 LSPQILVQRID--ISWNTRCMFLTKELGAPRDSIVKMVTKHPQLL 312
>TC221034 weakly similar to UP|O80618 (O80618) Predicted by genefinder and
genscan, partial (47%)
Length = 924
Score = 50.8 bits (120), Expect = 9e-07
Identities = 43/157 (27%), Positives = 73/157 (46%), Gaps = 8/157 (5%)
Frame = +3
Query: 154 VDFL-HSLGLTIEDIN----NYPLVLGCSVKKNMVPVLDYLGKLGVRKSTITQFLRTYPQ 208
+DFL H + + DI+ P +L SV + P L +L KLG T
Sbjct: 81 LDFLLHEVPIPYHDIHLSILRCPRLLVSSVNNRLRPTLHFLRKLGFNGPHSLTCQTTL-- 254
Query: 209 VLHASVVVDLVPVVKYLQGMDIKPDDIPRVLERYPEVLGFKLEGTMSTSVAYLIGIGVGR 268
+L +SV L+P +++L+G+ +++ ++ R P +L F +E + V + +
Sbjct: 255 LLVSSVEDTLLPKIEFLKGLGFTHEEVANMVVRSPGLLTFSVEKNLGPKVEFFL------ 416
Query: 269 RELGGI---LTRFPEILGMRVGRVIKPFVEYLESLGI 302
RE+ G L RFP+ + R IKP L +G+
Sbjct: 417 REMNGDVAELKRFPQYFSFSLERRIKPRFGMLRRVGV 527
>TC206953 similar to UP|O80618 (O80618) Predicted by genefinder and genscan,
partial (14%)
Length = 1027
Score = 48.9 bits (115), Expect = 4e-06
Identities = 35/127 (27%), Positives = 61/127 (47%)
Frame = +2
Query: 166 DINNYPLVLGCSVKKNMVPVLDYLGKLGVRKSTITQFLRTYPQVLHASVVVDLVPVVKYL 225
D+N P +L SVK + P L YL +LG + + + +L ++V L+P +K+L
Sbjct: 461 DVNKCPRLLTSSVKDQLRPCLVYLRRLGFKDLGALAYQDSV--LLVSNVENTLIPKLKFL 634
Query: 226 QGMDIKPDDIPRVLERYPEVLGFKLEGTMSTSVAYLIGIGVGRRELGGILTRFPEILGMR 285
+ + + D++ ++ R P +L F +E Y G +GR+ L FP+
Sbjct: 635 ETLGLSKDEVRSMVLRCPALLTFSIENNFQPKYEYFAG-EMGRKL--EELKEFPQYFAFS 805
Query: 286 VGRVIKP 292
+ IKP
Sbjct: 806 LENRIKP 826
>TC229367 similar to GB|AAP75804.1|32189299|BT009654 At4g09620 {Arabidopsis
thaliana;} , partial (68%)
Length = 1126
Score = 43.5 bits (101), Expect = 1e-04
Identities = 21/73 (28%), Positives = 47/73 (63%), Gaps = 1/73 (1%)
Frame = +1
Query: 288 RVIKPFVEYLESLGIPRLAIARLIETQPYILGFDLDEKVKPNVKSLEE-FNVRETSLASI 346
+ + ++YL SL + ++ L++ P +LG +L+E++K NVK LEE ++++ SL +
Sbjct: 475 KTVNGVLDYLRSLNLSDDDLSILLKKFPEVLGCNLEEELKGNVKILEEQWSIKGNSLRKL 654
Query: 347 IAQYPDIIGTDLE 359
+ + P ++G +++
Sbjct: 655 LLRNPKVLGYNVD 693
Score = 38.9 bits (89), Expect = 0.004
Identities = 23/73 (31%), Positives = 42/73 (57%), Gaps = 1/73 (1%)
Frame = +1
Query: 180 KNMVPVLDYLGKLGVRKSTITQFLRTYPQVLHASVVVDLVPVVKYLQGM-DIKPDDIPRV 238
K + VLDYL L + ++ L+ +P+VL ++ +L VK L+ IK + + ++
Sbjct: 475 KTVNGVLDYLRSLNLSDDDLSILLKKFPEVLGCNLEEELKGNVKILEEQWSIKGNSLRKL 654
Query: 239 LERYPEVLGFKLE 251
L R P+VLG+ ++
Sbjct: 655 LLRNPKVLGYNVD 693
Score = 38.5 bits (88), Expect = 0.005
Identities = 22/67 (32%), Positives = 38/67 (55%), Gaps = 1/67 (1%)
Frame = +1
Query: 221 VVKYLQGMDIKPDDIPRVLERYPEVLGFKLEGTMSTSVAYL-IGIGVGRRELGGILTRFP 279
V+ YL+ +++ DD+ +L+++PEVLG LE + +V L + L +L R P
Sbjct: 490 VLDYLRSLNLSDDDLSILLKKFPEVLGCNLEEELKGNVKILEEQWSIKGNSLRKLLLRNP 669
Query: 280 EILGMRV 286
++LG V
Sbjct: 670 KVLGYNV 690
Score = 36.2 bits (82), Expect = 0.024
Identities = 26/87 (29%), Positives = 46/87 (51%), Gaps = 1/87 (1%)
Frame = +1
Query: 240 ERYPEVLGFKLEGTMSTSVAYLIGIGVGRRELGGILTRFPEILGMRVGRVIKPFVEYL-E 298
ER EV K T++ + YL + + +L +L +FPE+LG + +K V+ L E
Sbjct: 448 ERSKEVPKLK---TVNGVLDYLRSLNLSDDDLSILLKKFPEVLGCNLEEELKGNVKILEE 618
Query: 299 SLGIPRLAIARLIETQPYILGFDLDEK 325
I ++ +L+ P +LG+++D K
Sbjct: 619 QWSIKGNSLRKLLLRNPKVLGYNVDCK 699
>TC234356 weakly similar to GB|AAN18117.1|23308295|BT000548
At4g38160/F20D10_280 {Arabidopsis thaliana;} , partial
(45%)
Length = 460
Score = 43.5 bits (101), Expect = 1e-04
Identities = 29/93 (31%), Positives = 47/93 (50%), Gaps = 4/93 (4%)
Frame = +3
Query: 155 DFLHSLGLTIEDIN----NYPLVLGCSVKKNMVPVLDYLGKLGVRKSTITQFLRTYPQVL 210
+FL S+GL+ D+ N+P +L V K +VP YL K G I + +P +L
Sbjct: 81 EFLKSIGLSEADLQAVAVNFPAILSRDVNKLLVPNYAYLKKRGFEDRQIVALVVGFPPIL 260
Query: 211 HASVVVDLVPVVKYLQGMDIKPDDIPRVLERYP 243
S+ L P +K+L +D+ + V++ YP
Sbjct: 261 IKSIQNSLEPRIKFL--VDVMGRQVDEVID-YP 350
>BE806270
Length = 420
Score = 30.8 bits (68), Expect = 0.99
Identities = 26/116 (22%), Positives = 52/116 (44%)
Frame = +3
Query: 218 LVPVVKYLQGMDIKPDDIPRVLERYPEVLGFKLEGTMSTSVAYLIGIGVGRRELGGILTR 277
LV ++ + D+ P D+PR L+ + T A+ + +L
Sbjct: 129 LVNLLLCSRKFDLNPLDLPRKLD------------LLKTRFAF------SAATVAKVLEG 254
Query: 278 FPEILGMRVGRVIKPFVEYLESLGIPRLAIARLIETQPYILGFDLDEKVKPNVKSL 333
FP++L + I V++L GIP I ++ P +LG ++++++P V+ +
Sbjct: 255 FPDVL-ITSETEITNVVDFLVEFGIPGDEIDLVVGLFPRVLGIGVEDRLRPLVREI 419
>AW200664 similar to GP|9971154|dbj mkpA {Dictyostelium discoideum}, partial
(0%)
Length = 399
Score = 29.6 bits (65), Expect = 2.2
Identities = 29/108 (26%), Positives = 44/108 (39%)
Frame = -3
Query: 49 LPCNYIRKIRFITKLQCSVAGRTDSYRTSAADSQGSKRDSGEIQRKRRGASSLYAGYARP 108
LP R R + +GR + + A+ + ++ S I+ ++S++ GY RP
Sbjct: 325 LPRTRKRHCRHRNRPLSKASGRGINGQYVFAEMRANEGQSSSIECLTSSSASVFGGYLRP 146
Query: 109 SLSEMKKDKATLRKVVYEFLRGIGIVPDELDGLELPVTVDVMKERVDF 156
S S A+ R + LRG G L L VT D V F
Sbjct: 145 SPS------ASFRSCL-RLLRGFGFGAITLSLHSLSVTYDFYNNYVSF 23
>TC205717 homologue to UP|O65463 (O65463) Protein kinase ADK1-like protein ,
partial (76%)
Length = 734
Score = 29.6 bits (65), Expect = 2.2
Identities = 14/41 (34%), Positives = 21/41 (51%)
Frame = +3
Query: 148 DVMKERVDFLHSLGLTIEDINNYPLVLGCSVKKNMVPVLDY 188
D + RV+++HS G DI ++G K N V +DY
Sbjct: 219 DQLINRVEYMHSRGFLHRDIKPDNFLMGLGRKANQVYAIDY 341
>TC234620 weakly similar to GB|AAO63444.1|28951041|BT005380 At1g62110
{Arabidopsis thaliana;} , partial (8%)
Length = 792
Score = 28.9 bits (63), Expect = 3.8
Identities = 17/54 (31%), Positives = 29/54 (53%), Gaps = 3/54 (5%)
Frame = +1
Query: 149 VMKERVDFLHSLGLTIEDINNYPLVLGCSVKKNMVP---VLDYLGKLGVRKSTI 199
+MK + + L EDI P +LGC+++K ++P V+ L G+ KS +
Sbjct: 466 IMKMLNFLVKDMSLPPEDIARCPEILGCNLEKTVIPRFAVVKNLKSRGLIKSDL 627
>TC227622 similar to UP|Q7XJT9 (Q7XJT9) NAM (No apical meristem)-like
protein, partial (61%)
Length = 1113
Score = 28.5 bits (62), Expect = 4.9
Identities = 13/23 (56%), Positives = 14/23 (60%)
Frame = +2
Query: 13 NFSGTITKPNFPFPPTEMPILTF 35
N SG IT P F F PTE +L F
Sbjct: 110 NMSGEITLPGFRFHPTEEELLDF 178
>TC228745 similar to UP|O65201 (O65201) Acyl-CoA oxidase , partial (68%)
Length = 1796
Score = 27.7 bits (60), Expect = 8.4
Identities = 10/18 (55%), Positives = 13/18 (71%)
Frame = -3
Query: 50 PCNYIRKIRFITKLQCSV 67
PCN IRKI+ I K C++
Sbjct: 1359 PCNVIRKIKCINKFPCNI 1306
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.141 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,355,728
Number of Sequences: 63676
Number of extensions: 165274
Number of successful extensions: 899
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 862
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 889
length of query: 359
length of database: 12,639,632
effective HSP length: 98
effective length of query: 261
effective length of database: 6,399,384
effective search space: 1670239224
effective search space used: 1670239224
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC122727.11


