

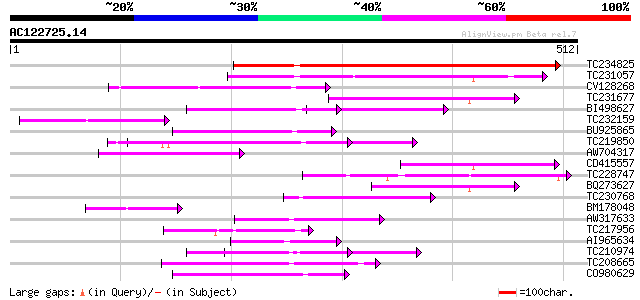
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122725.14 + phase: 0
(512 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC234825 468 e-132
TC231057 homologue to UP|Q9W1K2 (Q9W1K2) CG12491-PA, partial (8%) 127 9e-30
CV128268 111 7e-25
TC231677 similar to UP|O23677 (O23677) T7I23.8 protein, partial ... 105 5e-23
BI498627 94 2e-19
TC232159 92 6e-19
BU925865 similar to GP|20453085|gb At1g02150/T7I23.8 {Arabidopsi... 90 2e-18
TC219850 weakly similar to UP|Q9FKI5 (Q9FKI5) Emb|CAB69839.1, pa... 77 2e-14
AW704317 weakly similar to GP|6958202|gb| DNA-binding protein {T... 72 8e-13
CD415557 71 1e-12
TC228747 weakly similar to UP|Q9XI21 (Q9XI21) F9L1.43, partial (... 71 1e-12
BQ273627 70 2e-12
TC230768 69 5e-12
BM178048 61 1e-09
AW317633 similar to PIR|T46163|T46 nodulin / glutamate-ammonia l... 56 3e-08
TC217956 similar to UP|O65793 (O65793) Leaf protein, partial (30%) 56 3e-08
AI965634 56 4e-08
TC210974 similar to UP|Q6K9W7 (Q6K9W7) Pentatricopeptide (PPR) r... 55 6e-08
TC208665 weakly similar to UP|O65567 (O65567) Puative protein, p... 55 1e-07
CO980629 52 5e-07
>TC234825
Length = 874
Score = 468 bits (1205), Expect = e-132
Identities = 231/295 (78%), Positives = 264/295 (89%)
Frame = +1
Query: 203 TSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCL 262
TSPD+VTFNL L ACAS+NDVETAERVLL+LKKAK+DPDWVTYSTLTNLYI+NAS L
Sbjct: 1 TSPDIVTFNLWLAACASQNDVETAERVLLELKKAKIDPDWVTYSTLTNLYIKNAS----L 168
Query: 263 EKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCM 322
EKA +T+KEME RTSR+TRVAYSSLLSLH NMGN D+VNRIW KMKA F KM+D+EY+CM
Sbjct: 169 EKAGATVKEMENRTSRKTRVAYSSLLSLHTNMGNKDDVNRIWEKMKASFRKMNDNEYICM 348
Query: 323 ISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGV 382
ISSL+KLGDFAG E+LY+EWESVSGTNDVRVSN+LL SY++Q QMEMAE FCNQ+V+KGV
Sbjct: 349 ISSLLKLGDFAGAEDLYREWESVSGTNDVRVSNILLGSYINQDQMEMAEDFCNQIVQKGV 528
Query: 383 CLSYSSWELLTRGYLKKKDVKKFLHYFGKAISSVKQWIPDPRLVQEAFTVIQEQAHIEGA 442
Y++WEL T GYLK+KDV+KFL YF KAISSV +W PD RLVQEAF +I+EQAH +GA
Sbjct: 529 IPCYTTWELFTWGYLKRKDVEKFLDYFSKAISSVTKWSPDQRLVQEAFKIIEEQAHTKGA 708
Query: 443 EQLLVILRNAGHVNTNIYNLFLKTYAAAGKMPLVVAERMKKDNVQLDKETHRLLD 497
EQLLVILRNAGHVNTNIYNLFLKTYA AGKMP++VAERM+KDNV+LD+ET RLLD
Sbjct: 709 EQLLVILRNAGHVNTNIYNLFLKTYATAGKMPMIVAERMRKDNVKLDEETRRLLD 873
>TC231057 homologue to UP|Q9W1K2 (Q9W1K2) CG12491-PA, partial (8%)
Length = 1050
Score = 127 bits (320), Expect = 9e-30
Identities = 82/293 (27%), Positives = 154/293 (51%), Gaps = 4/293 (1%)
Frame = +3
Query: 197 EELKVNTSP-DVVTFNLLLTACASENDVETAERVLLQLKKAKVDP-DWVTYSTLTNLYIR 254
E +K T P T+++ + +CAS ND+ AERV ++K W TYS L ++Y++
Sbjct: 6 ELMKKRTIPMSPFTYHIWMNSCASSNDLGGAERVYEEMKTENEGQIGWHTYSNLASIYVK 185
Query: 255 NASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKM 314
EKA LK +E++ + R AY LL L+A GN+ EV+R+W +K+ +
Sbjct: 186 FKD----FEKAEMMLKMLEEQVKPKQRDAYHCLLGLYAGTGNLGEVHRVWDSLKSV-SPV 350
Query: 315 SDDEYVCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFC 374
++ Y+ M+S+L +L D G+ +KEWE+ + D R+ ++ ++++++Q +E AE+
Sbjct: 351 TNFSYLVMLSTLRRLNDMEGLTKCFKEWEASCVSYDARLVSVCVSAHLNQNMLEEAELVF 530
Query: 375 NQLVEKGVCLSYSSWELLTRGYLKKKDVKKFLHYFGKAISSVK--QWIPDPRLVQEAFTV 432
+ + + E + +LKK ++ + + A+S VK +W P P++V
Sbjct: 531 EEASRRSKGPFFRVREEFMKFFLKKHELDAAVRHLEAALSEVKGDKWRPSPQVVGAFLKY 710
Query: 433 IQEQAHIEGAEQLLVILRNAGHVNTNIYNLFLKTYAAAGKMPLVVAERMKKDN 485
+E+ ++G ++L IL+ N + ++K+ A K + MK+D+
Sbjct: 711 YEEETDVDGVDELSKILK-----ANNFDDSWIKSCITASKSSPEIDPGMKEDS 854
>CV128268
Length = 864
Score = 111 bits (278), Expect = 7e-25
Identities = 68/202 (33%), Positives = 113/202 (55%), Gaps = 2/202 (0%)
Frame = +2
Query: 90 EVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYV 149
+V EWM + + + D AVQLDLI +V G+ SAE++ + L D + ALL+ YV
Sbjct: 269 QVSEWMSSKG-LPISSRDQAVQLDLIGRVHGVESAERYLQSLSDGDKTWKVHGALLNCYV 445
Query: 150 QNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKVN-TSPDVV 208
+ L +K+ +LM KM + GF+ S + YN +MSLY + EKVP + E++K + P++
Sbjct: 446 REGLVDKSLSLMQKMKDMGFV-SFLNYNNIMSLYTQTQQYEKVPGVLEQMKKDGVPPNIF 622
Query: 209 TFNLLLTACASENDVETAERVLLQL-KKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAAS 267
++ + + + D+ E++L ++ ++ + DW+TYS +TN YI+ D
Sbjct: 623 SYRICINSYCVRGDLANVEKLLEEMEREPHIGIDWITYSMVTNFYIK----ADMRXXXLV 790
Query: 268 TLKEMEKRTSRETRVAYSSLLS 289
L + E T R VAY+ L+S
Sbjct: 791 CLMKCEXXTHRGNTVAYNHLIS 856
>TC231677 similar to UP|O23677 (O23677) T7I23.8 protein, partial (14%)
Length = 833
Score = 105 bits (262), Expect = 5e-23
Identities = 59/175 (33%), Positives = 93/175 (52%), Gaps = 3/175 (1%)
Frame = +2
Query: 289 SLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSGT 348
SL+ ++G DEV R+W K+ F + + Y +ISSLVKL D E LY+EW SV +
Sbjct: 2 SLYGSVGKKDEVCRVWNTYKSIFPSIPNLGYHAIISSLVKLDDIEVAEKLYEEWISVKSS 181
Query: 349 NDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKDVKKFLHY 408
D R+ NLL+ YV +G + A F Q++ G + ++WE+L+ G++ K + + L
Sbjct: 182 YDPRIGNLLMGWYVKKGDTDKALSFFEQMLNDGCIPNSNTWEILSEGHIADKRISEALSC 361
Query: 409 FGKAI---SSVKQWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAGHVNTNIY 460
+A K W P P + + QEQ +E AE L+ +LR + + +Y
Sbjct: 362 LKEAFMVAGGSKSWRPKPSYLSAFLELCQEQDDMESAEVLIGLLRQSKFNKSKVY 526
>BI498627
Length = 421
Score = 93.6 bits (231), Expect = 2e-19
Identities = 54/142 (38%), Positives = 81/142 (57%), Gaps = 2/142 (1%)
Frame = +3
Query: 160 LMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKV-NTSPDVVTFNLLLTACA 218
LM KM E S +PYN +M+LY G+ EK+P L +E+K N D T+N+ + A A
Sbjct: 6 LMEKMKELSLPLSSMPYNSLMTLYTKVGQPEKIPSLIQEMKASNVMLDSYTYNVWMRALA 185
Query: 219 SENDVETAERVLLQLKK-AKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTS 277
+ ND+ ERV ++K+ +V DW TYS L ++++ D KA LKE+EKR +
Sbjct: 186 AVNDISGVERVHDEMKRGGQVTGDWTTYSNLASIFVDAGLFD----KAEVALKELEKRNA 353
Query: 278 RETRVAYSSLLSLHANMGNVDE 299
+ AY L++L+ GN+ E
Sbjct: 354 FKDLTAYQFLITLYGRTGNLYE 419
Score = 43.9 bits (102), Expect = 2e-04
Identities = 27/129 (20%), Positives = 65/129 (49%), Gaps = 1/129 (0%)
Frame = +3
Query: 269 LKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVK 328
+++M++ + + + Y+SL++L+ +G +++ + +MKA + Y + +L
Sbjct: 9 MEKMKELSLPLSSMPYNSLMTLYTKVGQPEKIPSLIQEMKASNVMLDSYTYNVWMRALAA 188
Query: 329 LGDFAGVENLYKEWE-SVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYS 387
+ D +GVE ++ E + T D + L + +VD G + AE+ +L ++ +
Sbjct: 189 VNDISGVERVHDEMKRGGQVTGDWTTYSNLASIFVDAGLFDKAEVALKELEKRNAFKDLT 368
Query: 388 SWELLTRGY 396
+++ L Y
Sbjct: 369 AYQFLITLY 395
>TC232159
Length = 483
Score = 92.0 bits (227), Expect = 6e-19
Identities = 51/135 (37%), Positives = 76/135 (55%)
Frame = +1
Query: 10 LATTARHLSTEPARRGVTAASSSSGGDTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRK 69
L R +S+ + S D L R+ L PKRSA + KW +G+ +
Sbjct: 82 LLNRTRFVSSGAVSSDLVEESVEGVDDDLRSRIFRLRLPKRSATNVLQKWVLQGNPITLS 261
Query: 70 YQLNRMIRELRKNKRYKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFE 129
QL + +ELR+++RYKHALE+ EWM + +L + DYA ++DL TKV G+++AE++FE
Sbjct: 262 -QLRDISKELRRSQRYKHALEISEWMVSNEEYELSDSDYAARIDLTTKVFGIDAAERYFE 438
Query: 130 DLPDKMRGQPTCTAL 144
LP + T TAL
Sbjct: 439 GLPLATKTTETYTAL 483
>BU925865 similar to GP|20453085|gb At1g02150/T7I23.8 {Arabidopsis thaliana},
partial (27%)
Length = 443
Score = 90.1 bits (222), Expect = 2e-18
Identities = 52/150 (34%), Positives = 87/150 (57%), Gaps = 2/150 (1%)
Frame = +3
Query: 148 YVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEE-LKVNTSPD 206
YV + KAE+L M G++ +P+N MM+LY++ + KV L E ++ N D
Sbjct: 3 YVHSRSKEKAESLFDTMRSKGYVIHALPFNVMMTLYMNLNEYAKVDILASEMMEKNIQLD 182
Query: 207 VVTFNLLLTACASENDVETAERVLLQLKK-AKVDPDWVTYSTLTNLYIRNASVDDCLEKA 265
+ T+N+ L++C S+ VE E+V Q++K + P+W T+ST+ ++YIR D EKA
Sbjct: 183 IYTYNIWLSSCGSQGSVEKMEQVFEQMEKDPSIIPNWSTFSTMASMYIRM----DQNEKA 350
Query: 266 ASTLKEMEKRTSRETRVAYSSLLSLHANMG 295
L+++E R R+ + LLSL+ ++G
Sbjct: 351 EECLRKVEGRIKGRDRIPFHYLLSLYGSVG 440
Score = 29.6 bits (65), Expect = 3.4
Identities = 22/95 (23%), Positives = 43/95 (45%), Gaps = 3/95 (3%)
Frame = +3
Query: 97 LQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDL---PDKMRGQPTCTALLHAYVQNNL 153
++ +I+L Y + L + E+ FE + P + T + + Y++ +
Sbjct: 159 MEKNIQLDIYTYNIWLSSCGSQGSVEKMEQVFEQMEKDPSIIPNWSTFSTMASMYIRMDQ 338
Query: 154 TNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGK 188
KAE + K+ R +P++ ++SLY S GK
Sbjct: 339 NEKAEECLRKVEGRIKGRDRIPFHYLLSLYGSVGK 443
>TC219850 weakly similar to UP|Q9FKI5 (Q9FKI5) Emb|CAB69839.1, partial (53%)
Length = 1153
Score = 77.0 bits (188), Expect = 2e-14
Identities = 55/224 (24%), Positives = 112/224 (49%), Gaps = 3/224 (1%)
Frame = +2
Query: 89 LEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMR--GQPTCTALLH 146
LEV + M+ D+K+ + ++ +K RG ++A K FE+L K GQ T ++++
Sbjct: 188 LEVVKEME-DADVKVSDCILCTVVNGFSKKRGFSAAVKVFEELISKGNEPGQVTYASVIN 364
Query: 147 AYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKV-NTSP 205
AY + +KAE + +M + GF + Y+ M+ +Y G++ KL ++K P
Sbjct: 365 AYWRLGQYSKAEEVFLEMEQKGFDKCVYAYSTMIVMYGRTGRVRSAMKLVAKMKERGCKP 544
Query: 206 DVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKA 265
+V +N L+ + +++ E++ ++K+ +V PD V+Y+++ Y + + C++
Sbjct: 545 NVWIYNSLIDMHGRDKNLKQLEKLWKEMKRRRVAPDKVSYTSIIGAYSKAGEFETCVK-- 718
Query: 266 ASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKA 309
E R ++ + + +G VDE+ ++ MKA
Sbjct: 719 --LFNEYRMNGGLIDRAMAGIMVGVFSKVGQVDELVKLLQDMKA 844
Score = 69.7 bits (169), Expect = 3e-12
Identities = 57/269 (21%), Positives = 114/269 (42%), Gaps = 7/269 (2%)
Frame = +2
Query: 107 DYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCT------ALLHAYVQNNLTNKAEAL 160
+Y++ LI L + E+L + +G+ T L+H Y++ L K +
Sbjct: 20 EYSIYSKLIYSFASLGEVD-VAEELVREAKGKTTIKDPEVYLKLVHMYIEEGLLEKTLEV 196
Query: 161 MSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEEL-KVNTSPDVVTFNLLLTACAS 219
+ +M + S +++ + K+FEEL P VT+ ++ A
Sbjct: 197 VKEMEDADVKVSDCILCTVVNGFSKKRGFSAAVKVFEELISKGNEPGQVTYASVINAYWR 376
Query: 220 ENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRE 279
AE V L++++ D YST+ +Y R V ++ A +M++R +
Sbjct: 377 LGQYSKAEEVFLEMEQKGFDKCVYAYSTMIVMYGRTGRVRSAMKLVA----KMKERGCKP 544
Query: 280 TRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLY 339
Y+SL+ +H N+ ++ ++W +MK Y +I + K G+F L+
Sbjct: 545 NVWIYNSLIDMHGRDKNLKQLEKLWKEMKRRRVAPDKVSYTSIIGAYSKAGEFETCVKLF 724
Query: 340 KEWESVSGTNDVRVSNLLLTSYVDQGQME 368
E+ G D ++ +++ + GQ++
Sbjct: 725 NEYRMNGGLIDRAMAGIMVGVFSKVGQVD 811
>AW704317 weakly similar to GP|6958202|gb| DNA-binding protein {Triticum
aestivum}, partial (21%)
Length = 399
Score = 71.6 bits (174), Expect = 8e-13
Identities = 39/132 (29%), Positives = 65/132 (48%)
Frame = +2
Query: 81 KNKRYKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPT 140
+ K Y A ++ +W++ ++ +E DYA QLDLI K+RGL AEK+ E +P+ RG+
Sbjct: 2 RRKMYGRAFQLFQWLESNKKLEFMESDYASQLDLIAKLRGLPQAEKYIESVPESFRGELL 181
Query: 141 CTALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELK 200
LL N +E + +KM + + N+++ LY K + L K
Sbjct: 182 YRTLLANCASQNNLIASEKIFNKMKDLDLPLTVFACNQLLLLYKKLDKKKIADVLLLMEK 361
Query: 201 VNTSPDVVTFNL 212
N P + T+ +
Sbjct: 362 ENVKPSLFTYRI 397
>CD415557
Length = 539
Score = 71.2 bits (173), Expect = 1e-12
Identities = 39/145 (26%), Positives = 78/145 (52%), Gaps = 2/145 (1%)
Frame = -3
Query: 354 SNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKDVKKFLHYFGKAI 413
+ +LL +YV G ME AE ++KG C +Y + E+L GY+ + + + +A+
Sbjct: 537 TRVLLGAYVRNGLMEEAESLHLHTLQKGGCPNYKTLEILMEGYVNWPKMDEAIITMKRAL 358
Query: 414 SSVK--QWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAGHVNTNIYNLFLKTYAAAG 471
+ +K W P +V +++ ++E A++ + + N G V+ ++Y + L+ + +A
Sbjct: 357 AMMKDCHWRPPHGIVLALAEYLEKDGNLEYADKYITDIHNLGLVSLSLYKVLLRMHLSAN 178
Query: 472 KMPLVVAERMKKDNVQLDKETHRLL 496
K P + + M +D V++D ET +L
Sbjct: 177 KPPFHILKMMDEDKVEIDNETLSIL 103
>TC228747 weakly similar to UP|Q9XI21 (Q9XI21) F9L1.43, partial (35%)
Length = 924
Score = 71.2 bits (173), Expect = 1e-12
Identities = 62/251 (24%), Positives = 112/251 (43%), Gaps = 8/251 (3%)
Frame = +1
Query: 265 AASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMIS 324
A + LKEME +E + ++LL L+AN+G DEV RIW K C K ++ + +
Sbjct: 1 AEAMLKEMEGENLKENQWVCATLLRLYANLGKADEVERIW---KVCESKPRVEDCLAAVE 171
Query: 325 SLVKLGDFAGVENLY----KEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEK 380
+ KL E ++ K+W+ S V LL Y + + + + +
Sbjct: 172 AWGKLNKIEEAEAVFEMVSKKWKLNSKNYSV-----LLKIYANNKMLTKGKELVKLMADS 336
Query: 381 GVCLSYSSWELLTRGYLKKKDVKKFLHYFGKAISSVKQWIPDPRLVQEAFTVIQEQAHIE 440
GV + +W+ L + Y++ +V+K KAI Q P ++ +
Sbjct: 337 GVRIGPLTWDALVKLYIQAGEVEKADSILHKAIQQ-NQLQPMFTTYLAILEQYAKRGDVH 513
Query: 441 GAEQLLVILRNAGHVN-TNIYNLFLKTYAAAGKMPLVVAERMKKDNVQLDKETHR---LL 496
+E++ + +R AG+ + + + + ++ Y A + ER+K DN+ +K L+
Sbjct: 514 NSEKIFLKMRQAGYTSRISQFQVLIQAYVNAKVPAYGIRERIKADNLFPNKTLANQLALV 693
Query: 497 DLTSKMCVSDV 507
D K VSD+
Sbjct: 694 DAFRKNAVSDL 726
>BQ273627
Length = 420
Score = 70.5 bits (171), Expect = 2e-12
Identities = 39/137 (28%), Positives = 68/137 (49%), Gaps = 3/137 (2%)
Frame = -3
Query: 327 VKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSY 386
VKLG+ E + EWE T D RV N+LL Y +G +E AE ++V +G
Sbjct: 418 VKLGELDKAEKVLGEWELSGNTCDFRVPNILLIGYCQRGLVEKAEALLRKMVAEGKTPIP 239
Query: 387 SSWELLTRGYLKKKDVKKFLHYFGKAI---SSVKQWIPDPRLVQEAFTVIQEQAHIEGAE 443
+SW ++ GY+ K++++K +A+ + K+W P ++ F+ + IE AE
Sbjct: 238 NSWSIVASGYVAKENMEKAFQCMKEAVAVHAQNKRWRPKVDVISSIFSWVTNNRDIEEAE 59
Query: 444 QLLVILRNAGHVNTNIY 460
+ ++ +N +Y
Sbjct: 58 DFVNSWKSVNAMNRGMY 8
>TC230768
Length = 822
Score = 68.9 bits (167), Expect = 5e-12
Identities = 39/138 (28%), Positives = 74/138 (53%), Gaps = 1/138 (0%)
Frame = +2
Query: 248 LTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKM 307
L N+YI +D+ +++TL E EKR ++ + Y L+ L+ +G+ D++++IW +
Sbjct: 8 LANIYISVGHLDNA---SSNTLVETEKRITQRQWITYDFLIILYGGLGSKDKLDQIWNSL 178
Query: 308 KACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSGTN-DVRVSNLLLTSYVDQGQ 366
+ KM Y+C+ISS + LG V + +W+ + T+ D+ ++ ++ D G
Sbjct: 179 RMTKQKMISRNYICIISSYLMLGHTKEVGEVIDQWKQSTTTDFDMLACKKIMVAFRDMGL 358
Query: 367 MEMAEIFCNQLVEKGVCL 384
E+A L+EK + L
Sbjct: 359 AEIANNLNMILIEKNLSL 412
>BM178048
Length = 422
Score = 61.2 bits (147), Expect = 1e-09
Identities = 32/88 (36%), Positives = 53/88 (59%)
Frame = +2
Query: 69 KYQLNRMIRELRKNKRYKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFF 128
K+++ +++LR K Y+ AL++ E M +N IK V D+A+ LDL+ K RG+ +AE +F
Sbjct: 161 KWEVGDTLKKLRDRKLYQPALKLSETMAKRNMIKTVS-DHAIHLDLLAKARGITAAENYF 337
Query: 129 EDLPDKMRGQPTCTALLHAYVQNNLTNK 156
LP+ + ALL+ Y + +T K
Sbjct: 338 VSLPEPSKNHLCYGALLNCYCKELMTEK 421
>AW317633 similar to PIR|T46163|T46 nodulin / glutamate-ammonia ligase-like
protein - Arabidopsis thaliana, partial (26%)
Length = 484
Score = 56.2 bits (134), Expect = 3e-08
Identities = 31/135 (22%), Positives = 66/135 (47%)
Frame = +1
Query: 204 SPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLE 263
+P +VT+N ++ ++E ++ L++K V P+ +TY +L + Y S C++
Sbjct: 19 TPTIVTYNTVIEVFGKAGEIEKMDQHFLKMKHLGVKPNSITYCSLVSAY----SKVGCID 186
Query: 264 KAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMI 323
K S ++ +E ++ ++S + GN+ ++ ++ M+ C+ + + CMI
Sbjct: 187 KVDSIMRHVENSDVVLDTPFFNCIISAYGQAGNLKKMGELFLAMRERKCEPDNITFACMI 366
Query: 324 SSLVKLGDFAGVENL 338
S G V+NL
Sbjct: 367 QSYNTQGMTEAVQNL 411
>TC217956 similar to UP|O65793 (O65793) Leaf protein, partial (30%)
Length = 1224
Score = 56.2 bits (134), Expect = 3e-08
Identities = 40/139 (28%), Positives = 66/139 (46%), Gaps = 4/139 (2%)
Frame = +1
Query: 140 TCTALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYI----SNGKLEKVPKL 195
TC ++ Y + +A + +M E G +PV +N ++ Y+ +NG +++ L
Sbjct: 109 TCGIIISGYCKEGNMPEALRFLYRMKELGVDPNPVVFNSLIKGYLDTTDTNG-VDEALTL 285
Query: 196 FEELKVNTSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRN 255
EE + PDVVTF+ ++ A +S +E E + + KA ++PD YS L Y+R
Sbjct: 286 MEEFGIK--PDVVTFSTIMNAWSSAGLMENCEEIFNDMVKAGIEPDIHAYSILAKGYVRA 459
Query: 256 ASVDDCLEKAASTLKEMEK 274
KA L M K
Sbjct: 460 GQP----RKAEXLLTSMSK 504
Score = 38.1 bits (87), Expect = 0.009
Identities = 33/174 (18%), Positives = 71/174 (39%)
Frame = +1
Query: 208 VTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAAS 267
VT+N + A + E AER++L++ V P+ T + + Y + ++ + L
Sbjct: 1 VTYNTMARAYDQNGETERAERLILKMPYNIVKPNERTCGIIISGYCKEGNMPEALR---- 168
Query: 268 TLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLV 327
L M++ V ++SL+ + + + + V+ M+ K + ++++
Sbjct: 169 FLYRMKELGVDPNPVVFNSLIKGYLDTTDTNGVDEALTLMEEFGIKPDVVTFSTIMNAWS 348
Query: 328 KLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKG 381
G E ++ + D+ ++L YV GQ AE + + G
Sbjct: 349 SAGLMENCEEIFNDMVKAGIEPDIHAYSILAKGYVRAGQPRKAEXLLTSMSKYG 510
>AI965634
Length = 415
Score = 55.8 bits (133), Expect = 4e-08
Identities = 32/100 (32%), Positives = 55/100 (55%)
Frame = +2
Query: 200 KVNTSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVD 259
KV SP++VT+N+L+ +++A R+ QLK + + P VTY+TL I S
Sbjct: 17 KVGLSPNIVTYNILINGFCDVRKMDSAVRLFNQLKSSGLSPTLVTYNTL----IAGYSKV 184
Query: 260 DCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDE 299
+ L A +KEME+R ++V Y+ L+ A + + ++
Sbjct: 185 ENLAGALDLVKEMEERCIAPSKVTYTILIDAFARLNHTEK 304
>TC210974 similar to UP|Q6K9W7 (Q6K9W7) Pentatricopeptide (PPR)
repeat-containing protein-like, partial (3%)
Length = 637
Score = 55.5 bits (132), Expect = 6e-08
Identities = 46/180 (25%), Positives = 86/180 (47%), Gaps = 2/180 (1%)
Frame = +2
Query: 195 LFEE-LKVNTSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYI 253
+FEE L P +N+LL A + VE A+ V +++ + PD +Y+T+ + YI
Sbjct: 2 VFEEMLDAGIRPTRKAYNILLDAFSISGMVEQAQTVFKSMRRDRYFPDLCSYTTMLSAYI 181
Query: 254 RNASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCK 313
+ DD +E A K + + V Y +L+ +A + +++ V + + +M K
Sbjct: 182 ---NADD-MEGAEKFFKRLIQDGFEPNVVTYGTLIKGYAKINDLEMVMKKYEEMLMRGIK 349
Query: 314 MSDDEYVCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLT-SYVDQGQMEMAEI 372
+ ++ + K GDF + +KE ES D + N+LL+ + D+ + E E+
Sbjct: 350 ANQTILTTIMDAYGKSGDFDSAVHWFKEMESNGIPPDQKAKNVLLSLAKTDEEREEANEL 529
Score = 50.1 bits (118), Expect = 2e-06
Identities = 33/151 (21%), Positives = 76/151 (49%), Gaps = 1/151 (0%)
Frame = +2
Query: 160 LMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKVNTS-PDVVTFNLLLTACA 218
+ +M + G + YN ++ + +G +E+ +F+ ++ + PD+ ++ +L+A
Sbjct: 2 VFEEMLDAGIRPTRKAYNILLDAFSISGMVEQAQTVFKSMRRDRYFPDLCSYTTMLSAYI 181
Query: 219 SENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSR 278
+ +D+E AE+ +L + +P+ VTY TL Y A ++D LE +EM R +
Sbjct: 182 NADDMEGAEKFFKRLIQDGFEPNVVTYGTLIKGY---AKIND-LEMVMKKYEEMLMRGIK 349
Query: 279 ETRVAYSSLLSLHANMGNVDEVNRIWGKMKA 309
+ ++++ + G+ D + +M++
Sbjct: 350 ANQTILTTIMDAYGKSGDFDSAVHWFKEMES 442
>TC208665 weakly similar to UP|O65567 (O65567) Puative protein, partial (36%)
Length = 1204
Score = 54.7 bits (130), Expect = 1e-07
Identities = 39/198 (19%), Positives = 85/198 (42%)
Frame = +1
Query: 138 QPTCTALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFE 197
Q +L+ Q ++ L +M + GF S + +N M+ ++ KV +L+
Sbjct: 598 QELYNCVLNCCAQALPVDELSRLFDEMVQHGFAPSTITFNVMLDVFGKAKLFNKVWRLYC 777
Query: 198 ELKVNTSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNAS 257
K DV+T+N ++ A D + +++ Y+++ + Y
Sbjct: 778 MAKKQGLVDVITYNTIIAAYGKNKDFNNMSSTVQKMEFDGFSVSLEAYNSMLDAY----G 945
Query: 258 VDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDD 317
D +E S L++M+ Y++L++++ G ++EV + ++K C +
Sbjct: 946 KDGQMETFRSVLQKMKDSNCASDHYTYNTLINIYGEQGWINEVANVLTELKECGLRPD-- 1119
Query: 318 EYVCMISSLVKLGDFAGV 335
+C ++L+K AG+
Sbjct: 1120--LCSYNTLIKAYGIAGM 1167
>CO980629
Length = 541
Score = 52.4 bits (124), Expect = 5e-07
Identities = 36/161 (22%), Positives = 73/161 (44%), Gaps = 1/161 (0%)
Frame = -2
Query: 148 YVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEEL-KVNTSPD 206
+V+ T K + ++M G + YN ++ + + LE+ K+ + K +P+
Sbjct: 537 HVKAGRTEKVLKVYNQMKRLGCPADTISYNFIIESHCRDENLEEAAKILNLMVKKGVAPN 358
Query: 207 VVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAA 266
TFN + A +DV A R+ ++K+ P+ +TY+ L + + S D L+
Sbjct: 357 ASTFNFIFGCIAKLHDVNGAHRMYARMKELNCQPNTLTYNILMRMXXESRSTDMVLKMK- 181
Query: 267 STLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKM 307
KEM++ Y L+S+ +M + + ++ +M
Sbjct: 180 ---KEMDESQVEPNVNTYRILISMFCDMKHWNNAYKLMMEM 67
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.132 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,343,221
Number of Sequences: 63676
Number of extensions: 285924
Number of successful extensions: 1484
Number of sequences better than 10.0: 117
Number of HSP's better than 10.0 without gapping: 1402
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1445
length of query: 512
length of database: 12,639,632
effective HSP length: 101
effective length of query: 411
effective length of database: 6,208,356
effective search space: 2551634316
effective search space used: 2551634316
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122725.14


