

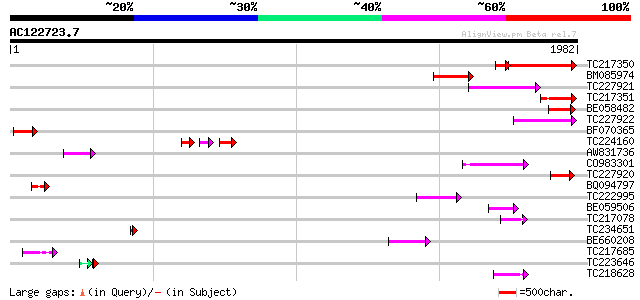
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122723.7 + phase: 0 /pseudo
(1982 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC217350 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like pro... 333 e-108
BM085974 homologue to PIR|T49914|T4 callose synthase catalytic s... 216 8e-56
TC227921 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like pro... 178 2e-44
TC217351 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like pro... 161 3e-39
BE058482 145 1e-34
TC227922 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like pro... 122 2e-27
BF070365 similar to PIR|T49914|T49 callose synthase catalytic su... 119 1e-26
TC224160 60 8e-19
AW831736 93 1e-18
CO983301 91 7e-18
TC227920 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like pro... 87 1e-16
BQ094797 76 1e-13
TC222995 weakly similar to UP|GLS2_YEAST (P40989) 1,3-beta-gluca... 75 4e-13
BE059506 similar to PIR|F86200|F8 protein F12K11.17 [imported] -... 64 7e-10
TC217078 59 2e-08
TC234651 54 5e-07
BE660208 52 3e-06
TC217685 similar to GB|AAM16165.1|20334818|AY094009 AT4g26750/F1... 49 2e-05
TC223646 43 2e-05
TC218628 similar to UP|Q9LWU7 (Q9LWU7) ESTs AU033035(S1515), par... 44 5e-04
>TC217350 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like protein
(Fragment), partial (31%)
Length = 1049
Score = 333 bits (855), Expect(2) = e-108
Identities = 185/237 (78%), Positives = 196/237 (82%)
Frame = +3
Query: 1745 KKLLMIGLIGINGLVTEEV*VYRLKKVGNLGGKKNKIIYSIQESGES*WRYCCHCDFLST 1804
KKLLMIG IGINGLV E V*VYRLKKVGNLGGK+NK I +I E GE *WRYC H DFLST
Sbjct: 141 KKLLMIGPIGINGLVIEGV*VYRLKKVGNLGGKRNKSISNILECGEL*WRYCYH*DFLST 320
Query: 1805 NMVLYIT*ILQRRAQKVFWCMAFRGW*SL*YYL**RLYLLGGVNSVQIFSLSSD*SRG*Y 1864
+MV YIT*IL+R+AQKVFW MAF GW* *Y L**R YLLGGVNSVQIF+LSSD*SR *Y
Sbjct: 321 SMVWYIT*ILRRKAQKVFWSMAFHGW*YS*YCL**RRYLLGGVNSVQIFNLSSD*SRE*Y 500
Query: 1865 S*LLLRFWSF*SPFHI*HRKTLLFAFLPSCQLVGECYRLRKH*SL*YVVLDFGDL*KLLL 1924
S LL RF SF* F I* +TLLFAFL SCQLVGEC RL KH*+L*Y L FGD *KLLL
Sbjct: 501 SSLLSRFLSF*LLFRI*QCRTLLFAFLLSCQLVGECCRLLKH*NL*YAGLGFGDQ*KLLL 680
Query: 1925 VVMR*SWVCFCSLLLPFWLGFLLFQNFKPECCSTKRSAEVCKFLVFLEDRGRSALLA 1981
VMR SWVC+CSLLL FWLGF LFQNFKP CCSTK SAEVCKFLVFL +GR+ALLA
Sbjct: 681 EVMRLSWVCYCSLLLLFWLGFHLFQNFKPVCCSTKHSAEVCKFLVFLVAKGRNALLA 851
Score = 79.7 bits (195), Expect(2) = e-108
Identities = 38/50 (76%), Positives = 41/50 (82%)
Frame = +2
Query: 1699 YLLYTKFLVIHTEVLLHIY*LLYQCGSWWEPGCLLHSCSTPLGLNGKKLL 1748
YLLY KFLVI TEVLLHI *L +QCG WW PGCLL SCS LGLNGKK++
Sbjct: 2 YLLYMKFLVILTEVLLHIS*LPHQCGLWWVPGCLLPSCSILLGLNGKKIV 151
>BM085974 homologue to PIR|T49914|T4 callose synthase catalytic subunit-like
protein - Arabidopsis thaliana, partial (7%)
Length = 427
Score = 216 bits (550), Expect = 8e-56
Identities = 115/140 (82%), Positives = 118/140 (84%)
Frame = +2
Query: 1480 ASTLHSVKEVLLIMSTYKLGREEMLVSTKFRCLKQK*LMEMESRH*VETCTGLGTVLISS 1539
ASTLHSVKE+LL MSTYK GREEM VS +F CLKQK*LM MES+H*VET T L TVL SS
Sbjct: 8 ASTLHSVKEMLLTMSTYK*GREEMWVSIRFLCLKQK*LMGMESKH*VETYTDLDTVLTSS 187
Query: 1540 ECCPVILLQLDFISALL*LFSLYTYFSMVASILFSVALKKG*VHKKPFETISLFKWLLLL 1599
ECCPVI QLDFISALL*LFSLY Y SMVASILFSV KK * KKPFETISLFKWLLLL
Sbjct: 188 ECCPVISPQLDFISALL*LFSLYMYSSMVASILFSVGSKKV*ALKKPFETISLFKWLLLL 367
Query: 1600 SHSFKLGF*WPCPC*WKLAW 1619
SH FKLGF*WPCPC*WK W
Sbjct: 368 SHLFKLGF*WPCPC*WKSVW 427
>TC227921 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like protein
(Fragment), partial (27%)
Length = 759
Score = 178 bits (451), Expect = 2e-44
Identities = 123/250 (49%), Positives = 143/250 (57%)
Frame = +1
Query: 1605 LGF*WPCPC*WKLAWKGASELHLVNLY*CSCSLLLYSSRFP*GPRLTILEGHCFMEVQNI 1664
L F PC C*WKLAW+G VNLY*C+ S LLY S F + TI+EGHCFMEVQ+
Sbjct: 1 LDFC*PCLC*WKLAWRGDFVRL*VNLY*CNYSWLLYFSHFLLEQKHTIMEGHCFMEVQST 180
Query: 1665 DPRVEDLWFSMPSLLIIIDFIREVTLSRVLSSWYYLLYTKFLVIHTEVLLHIY*LLYQCG 1724
V L MP+LLI I FI L + L+ + L T +L ++TE L I+*L QCG
Sbjct: 181 KALVAVLLCFMPNLLITIGFIPAAILLKGLN**FCLWSTIYLAMNTEECLPIF*LP*QCG 360
Query: 1725 SWWEPGCLLHSCSTPLGLNGKKLLMIGLIGINGLVTEEV*VYRLKKVGNLGGKKNKIIYS 1784
SW G LLHS S L+G++ LM IG NG T E VY LKKVGN GGK+N I
Sbjct: 361 SW*GHGFLLHSFSIHQVLSGRR*LMTTQIGKNG*ATVEALVYLLKKVGNPGGKRNMSISG 540
Query: 1785 IQESGES*WRYCCHCDFLSTNMVLYIT*ILQRRAQKVFWCMAFRGW*SL*YYL**RLYLL 1844
S S R F NM YIT R KVFW M F G *SL*Y++**R+YLL
Sbjct: 541 TLASAVSLLR*Y*PAAFSFINMAWYITFQSLMRRLKVFWFMVFHG**SL*YWV**RVYLL 720
Query: 1845 GGVNSVQIFS 1854
G SVQ S
Sbjct: 721 AGAASVQTTS 750
>TC217351 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like protein
(Fragment), partial (13%)
Length = 601
Score = 161 bits (407), Expect = 3e-39
Identities = 91/124 (73%), Positives = 95/124 (76%)
Frame = +1
Query: 1857 SD*SRG*YS*LLLRFWSF*SPFHI*HRKTLLFAFLPSCQLVGECYRLRKH*SL*YVVLDF 1916
SD*SR *Y LL RF SF H+ TLLFAF SCQLVGEC RL KH*+L*Y L F
Sbjct: 1 SD*SRE*YXXLLSRFLSFDCSXHM-XXWTLLFAF*LSCQLVGECCRLLKH*NL*YAGLGF 177
Query: 1917 GDL*KLLLVVMR*SWVCFCSLLLPFWLGFLLFQNFKPECCSTKRSAEVCKFLVFLEDRGR 1976
GD *KLLL VMR SWVC+C LLL FWLGFLLFQNFKP CCSTK SAEVCKFLVFL +GR
Sbjct: 178 GDQ*KLLLEVMRLSWVCYCLLLLLFWLGFLLFQNFKPVCCSTKHSAEVCKFLVFLVAKGR 357
Query: 1977 SALL 1980
SALL
Sbjct: 358 SALL 369
>BE058482
Length = 430
Score = 145 bits (367), Expect = 1e-34
Identities = 74/93 (79%), Positives = 78/93 (83%)
Frame = +2
Query: 1885 TLLFAFLPSCQLVGECYRLRKH*SL*YVVLDFGDL*KLLLVVMR*SWVCFCSLLLPFWLG 1944
TLLFAFL SCQLVGEC RL KH*+L*Y L FGD *KLLL VMR SWVC+CSLLL FWLG
Sbjct: 152 TLLFAFLLSCQLVGECCRLLKH*NL*YAGLGFGDQ*KLLLEVMRLSWVCYCSLLLLFWLG 331
Query: 1945 FLLFQNFKPECCSTKRSAEVCKFLVFLEDRGRS 1977
F LFQNFKP CCSTK SAEVCKFLVFL +GR+
Sbjct: 332 FHLFQNFKPVCCSTKHSAEVCKFLVFLVAKGRN 430
>TC227922 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like protein
(Fragment), partial (23%)
Length = 1119
Score = 122 bits (306), Expect = 2e-27
Identities = 103/219 (47%), Positives = 117/219 (53%)
Frame = +3
Query: 1762 EV*VYRLKKVGNLGGKKNKIIYSIQESGES*WRYCCHCDFLSTNMVLYIT*ILQRRAQKV 1821
E VY LKK GN GGK+N I ES S R F NM L+IT R KV
Sbjct: 3 EALVYLLKKFGNPGGKRNMSISGTPESVVSLLR*YYPSAFSFINMALFITFQSLIRL-KV 179
Query: 1822 FWCMAFRGW*SL*YYL**RLYLLGGVNSVQIFSLSSD*SRG*YS*LLLRFWSF*SPFHI* 1881
FW M F G *SL*Y++**R+YLL G SVQ + D*S +S* L * *
Sbjct: 180 FWFMVFLG**SL*YWV**RVYLLAGAASVQTTNFYFD*SWVLFS*HSLPSS*S**QLPR* 359
Query: 1882 HRKTLLFAFLPSCQLVGECYRLRKH*SL*YVVLDFGDL*KLLLVVMR*SWVCFCSLLLPF 1941
+T AFL SCQ GE L K SL L FG L + L M+* W CF S L
Sbjct: 360 QLRT**SAFLQSCQRAGEFC*LHKLASLSLKKLGFGGLFEHLHADMK**WACFSSHQLLS 539
Query: 1942 WLGFLLFQNFKPECCSTKRSAEVCKFLVFLEDRGRSALL 1980
W G LFQ+FK EC STK AEVC+FLVFL D+ +A L
Sbjct: 540 WHGSHLFQSFKLECFSTKLLAEVCRFLVFLVDKVMNAPL 656
>BF070365 similar to PIR|T49914|T49 callose synthase catalytic subunit-like
protein - Arabidopsis thaliana, partial (3%)
Length = 259
Score = 119 bits (299), Expect = 1e-26
Identities = 59/84 (70%), Positives = 66/84 (78%)
Frame = +2
Query: 14 PPPRRLVRTQTAGNLGESIFDSEVVPSSLVEIAPILRVANEVEKTHPRVAYLCRFYAFEK 73
P P+ ++RTQTAGNLGES+ D EVVP +VEIAPILRV +EVEKTHP YLCR YA EK
Sbjct: 8 PSPKLIMRTQTAGNLGESVIDREVVPYYMVEIAPILRVTDEVEKTHPMDDYLCRIYASEK 187
Query: 74 AHRLDPTSSGRGVRQFKTALLQRL 97
AH +DP SGRGVRQF TALL L
Sbjct: 188AHMVDPNISGRGVRQFTTALLHAL 259
>TC224160
Length = 932
Score = 60.5 bits (145), Expect(3) = 8e-19
Identities = 26/59 (44%), Positives = 40/59 (67%)
Frame = +2
Query: 734 CCSCSLGSNYSGILYGYPNLVCNILNIIWWDLWCIPSTGGDTYTRNA*VSVSVIAWCLQ 792
C SC+LGS G+ +G+ NLVC + N + W + S+ DT++ +A ++VSVIAWC+Q
Sbjct: 569 CSSCTLGSCTFGLFHGHSNLVCTLHNFVRRSCWSL*SSRRDTHSEHAKITVSVIAWCIQ 745
Score = 44.3 bits (103), Expect(3) = 8e-19
Identities = 18/49 (36%), Positives = 31/49 (62%), Gaps = 4/49 (8%)
Frame = +1
Query: 602 LFIV----AVVVYLSPNMLAAIFFMFPFIRRYLERSNYRIVMLMMWWSQ 646
LFI+ + +++ S A+ F+FP +RR++E S++ IV +WWSQ
Sbjct: 211 LFIILHAGSCIIFASEFTSTAVLFLFPMLRRWIENSDWHIVRFFLWWSQ 357
Score = 28.9 bits (63), Expect(3) = 8e-19
Identities = 20/51 (39%), Positives = 25/51 (48%)
Frame = +3
Query: 662 ASPLCW*GNA*EYIFPFQVHGVLGPPFIYKVGFQLLYRDKTSGWTYKGYYE 712
A LCW NA* I +V+ +L F + QL DKTS KG+ E
Sbjct: 357 AKHLCWKRNA**SICSHEVNYILVVAFDM*IFIQLFCPDKTSS*ANKGHNE 509
>AW831736
Length = 396
Score = 92.8 bits (229), Expect = 1e-18
Identities = 49/117 (41%), Positives = 71/117 (59%), Gaps = 3/117 (2%)
Frame = +2
Query: 187 LVPFNILPLDPDSANQAIMKFPEIQAAVYALRNTRGLP-WPNDYK--KKKDEDILDWLGS 243
L P+NI+PL+ S I FPE++AA+ A+R T P P +K ++D D+ D L
Sbjct: 44 LTPYNIIPLEAPSLTNPIRIFPEVKAAISAIRYTDQFPRLPAGFKISGQRDADMFDLLEF 223
Query: 244 MFGFQKHNVANQREHLILLLANVHIRQFPNPDQQPKLDECALTEVMKKLFKNYKKWC 300
+FGFQK NV NQRE+++L++AN R + PK+DE + EV K+ NY +WC
Sbjct: 224 VFGFQKDNVRNQRENVVLMIANKQSRLGIPAETDPKIDEKTINEVFLKVLDNYIRWC 394
>CO983301
Length = 825
Score = 90.5 bits (223), Expect = 7e-18
Identities = 82/228 (35%), Positives = 99/228 (42%)
Frame = -3
Query: 1584 KKPFETISLFKWLLLLSHSFKLGF*WPCPC*WKLAWKGASELHLVNLY*CSCSLLLYSSR 1643
K+P++ LF W + + C WKLA + L + L CSCS L S
Sbjct: 703 KRPWQLSLLFSW----------AYCFCCQWLWKLA*REDFALPWLILSSCSCS*PLCSLL 554
Query: 1644 FP*GPRLTILEGHCFMEVQNIDPRVEDLWFSMPSLLIIIDFIREVTLSRVLSSWYYLLYT 1703
+ I+E H +M NID +V D SM SLLI EV L R S Y L+
Sbjct: 553 SSLEQKHIIMEEHSYMGALNIDQQVVD*LSSM*SLLITTGCTHEVIL*RA*RSSCY*LFM 374
Query: 1704 KFLVIHTEVLLHIY*LLYQCGSWWEPGCLLHSCSTPLGLNGKKLLMIGLIGINGLVTEEV 1763
+ H + IY Q G W PGCLL SC L L K MIG IG G V
Sbjct: 373 NSMESHIAARIFIYSS*SQSGFWPRPGCLLLSCLILLALIC*KQWMIGQIGSGGWDILMV 194
Query: 1764 *VYRLKKVGNLGGKKNKIIYSIQESGES*WRYCCHCDFLSTNMVLYIT 1811
V+ L + GNLGG + Q SG * R H L TNM L T
Sbjct: 193 LVFHLIEAGNLGGMNKMNT*NTQISGVK*LR*FLHFXXLCTNMELSTT 50
>TC227920 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like protein
(Fragment), partial (13%)
Length = 675
Score = 86.7 bits (213), Expect = 1e-16
Identities = 50/86 (58%), Positives = 55/86 (63%)
Frame = +2
Query: 1889 AFLPSCQLVGECYRLRKH*SL*YVVLDFGDL*KLLLVVMR*SWVCFCSLLLPFWLGFLLF 1948
AFL SCQ GEC L K SL L FG L + LL M+* W CF S L W G LF
Sbjct: 110 AFLQSCQQAGECC*LHKLASLSLKKLGFGGLFEHLLADMK**WACFSSHQLLSWHGSHLF 289
Query: 1949 QNFKPECCSTKRSAEVCKFLVFLEDR 1974
Q+FKPEC STK SAEVC+FLVFL D+
Sbjct: 290 QSFKPECFSTKLSAEVCRFLVFLVDK 367
>BQ094797
Length = 423
Score = 76.3 bits (186), Expect = 1e-13
Identities = 44/63 (69%), Positives = 46/63 (72%)
Frame = +1
Query: 76 RLDPTSSGRGVRQFKTALLQRLERENDPTLKGRVKKSDAREMQSFYQHYYKKYIQALQNA 135
RLD SSG GV QFKTALL RLE EN T R K SDAREMQ+FY YY+KYIQAL A
Sbjct: 238 RLDLHSSGSGVCQFKTALLWRLENEN-LTTHVRRKTSDAREMQTFYGQYYEKYIQALDKA 414
Query: 136 ADK 138
ADK
Sbjct: 415 ADK 423
>TC222995 weakly similar to UP|GLS2_YEAST (P40989) 1,3-beta-glucan synthase
component GLS2 (1,3-beta-D-glucan-UDP
glucosyltransferase) , partial (3%)
Length = 490
Score = 74.7 bits (182), Expect = 4e-13
Identities = 65/159 (40%), Positives = 81/159 (50%)
Frame = +2
Query: 1421 LCQIKRQVS*QLVRDCWPIP*GFVSTMGILMSLIGFFT*QEGVSVKPPRLSI*VKIFLLA 1480
LC IK+ V * + I * TMGILM LI + T* VS + P LS VKI +
Sbjct: 14 LCPIKKLVL*L*GSEF*QIH*RSACTMGILMFLIEYST*PVVVSARLPVLSTLVKISIQD 193
Query: 1481 STLHSVKEVLLIMSTYKLGREEMLVSTKFRCLKQK*LMEMESRH*VETCTGLGTVLISSE 1540
S VKE+L IM+ ++L REEML S + LK+K L+ ++ T L + LISS
Sbjct: 194 SIQPYVKEILHIMNIFRLEREEMLGSIRLHYLKEKFLVVTVNKFSAVMYTDLDSFLISSG 373
Query: 1541 CCPVILLQLDFISALL*LFSLYTYFSMVASILFSVALKK 1579
CC IL QLD * LY F M I + L K
Sbjct: 374 CCLSILQQLDTTFVPC*QC*LYMPFYMGKHIWHCLVLVK 490
>BE059506 similar to PIR|F86200|F8 protein F12K11.17 [imported] - Arabidopsis
thaliana, partial (5%)
Length = 320
Score = 63.9 bits (154), Expect = 7e-10
Identities = 47/103 (45%), Positives = 51/103 (48%)
Frame = +2
Query: 1674 SMPSLLIIIDFIREVTLSRVLSSWYYLLYTKFLVIHTEVLLHIY*LLYQCGSWWEPGCLL 1733
SM SLLI EV L R YY L+ K + H V + IY QCG GCLL
Sbjct: 8 SMRSLLITTGCTHEVIL*RA*RYSYY*LFMKSMGAHIAVHIFIYSSQSQCGL*PLTGCLL 187
Query: 1734 HSCSTPLGLNGKKLLMIGLIGINGLVTEEV*VYRLKKVGNLGG 1776
SC L L GKK MIG IG G V V+ L K GNLGG
Sbjct: 188 LSCLILLALMGKKQWMIGQIGSGGWEIGVVLVFYLIKAGNLGG 316
>TC217078
Length = 1010
Score = 58.9 bits (141), Expect = 2e-08
Identities = 43/94 (45%), Positives = 49/94 (51%)
Frame = +1
Query: 1715 HIY*LLYQCGSWWEPGCLLHSCSTPLGLNGKKLLMIGLIGINGLVTEEV*VYRLKKVGNL 1774
HI+ + G W PGCLLH ST L L+GK+ I IG G TE V R KK G L
Sbjct: 7 HIFF*V*AVGLWLYPGCLLHIYSTRLDLSGKRWWKISEIGQIGYSTEVELVLREKKAGKL 186
Query: 1775 GGKKNKIIYSIQESGES*WRYCCHCDFLSTNMVL 1808
GGKKN +I G * + DF S NMVL
Sbjct: 187 GGKKNWLISGAWVQG--*QKPF*VLDFSSFNMVL 282
Score = 33.9 bits (76), Expect = 0.73
Identities = 33/89 (37%), Positives = 43/89 (48%)
Frame = +1
Query: 1882 HRKTLLFAFLPSCQLVGECYRLRKH*SL*YVVLDFGDL*KLLLVVMR*SWVCFCSLLLPF 1941
+R LL +L S LVGE +L S * D+G+ LL M W C LP
Sbjct: 499 YRTYLLLCWL-SFLLVGEFCQLLLPGSR**RGSDYGNPFDQLLASMMLVWECSSLYPLPS 675
Query: 1942 WLGFLLFQNFKPECCSTKRSAEVCKFLVF 1970
+ G L+Q+ K C T+ AE KFL+F
Sbjct: 676 FRGSHLYQHSKLA*CLTRLLAEG*KFLLF 762
>TC234651
Length = 601
Score = 54.3 bits (129), Expect = 5e-07
Identities = 22/24 (91%), Positives = 24/24 (99%)
Frame = +2
Query: 423 GWPMRADADFFCLPAERVVFDKSN 446
GWPMRADADFFCLPAE++VFDKSN
Sbjct: 44 GWPMRADADFFCLPAEKLVFDKSN 115
>BE660208
Length = 720
Score = 52.0 bits (123), Expect = 3e-06
Identities = 55/150 (36%), Positives = 74/150 (48%), Gaps = 4/150 (2%)
Frame = +3
Query: 1325 LQSLSRIW----TRSYIK*SFLVQRF*VRVSPKIKITP*FSHVEKVCKQ*I*TRITTWKK 1380
+QSLS++ R YI +FL + S K K P S V K+ K * R KK
Sbjct: 117 IQSLSKLI*MEKIRKYILLNFLEILSLGKESLKTKTMPSSSLVVKLFKLLT*IRTIILKK 296
Query: 1381 P*K*EICCKNFLRSMMV*GSQVFLD*ESIYLLEVFLPLHGLCQIKRQVS*QLVRDCWPIP 1440
*K* I ++ +R +M Q FL+* +++L EV LP GLCQIK+ * + I
Sbjct: 297 Q*K*GIFWRSSMR-IMAFALQAFLE*GNMFLQEVSLPWLGLCQIKKLAL*PWPNEF*QIL 473
Query: 1441 *GFVSTMGILMSLIGFFT*QEGVSVKPPRL 1470
F TM M L +FT* E V+ R+
Sbjct: 474 SKFACTMDTQMYLTEYFT*LEEALVRHHRV 563
>TC217685 similar to GB|AAM16165.1|20334818|AY094009 AT4g26750/F10M23_90
{Arabidopsis thaliana;} , partial (57%)
Length = 1586
Score = 49.3 bits (116), Expect = 2e-05
Identities = 33/126 (26%), Positives = 57/126 (45%), Gaps = 5/126 (3%)
Frame = +3
Query: 45 IAPILRVANEVEKTHPRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENDPT 104
+ P L+ A+E++K P VAY CR YA E+ ++ + + +L+++LE++
Sbjct: 69 LLPYLQRADELQKHEPLVAYYCRLYAMERGLKIPQSERTKTTNALLVSLMKQLEKDKKSI 248
Query: 105 LKGRVKKSDAREMQSFYQHYYKKYIQALQNAADKADRA-----QLTKAYQTANVLFEVLK 159
G D ++ F + + K ADK DRA K + A++ FE+L
Sbjct: 249 QLG---PEDNLYLEGFALNVFGK--------ADKQDRAGRADLNTAKTFYAASIFFEILN 395
Query: 160 AVNMTQ 165
Q
Sbjct: 396 QFGAVQ 413
>TC223646
Length = 526
Score = 42.7 bits (99), Expect(2) = 2e-05
Identities = 18/23 (78%), Positives = 19/23 (82%)
Frame = +1
Query: 288 VMKKLFKNYKKWCKYLDRKSSLW 310
V KKLF+NYKKWC YL KSSLW
Sbjct: 97 VKKKLFRNYKKWC*YLGCKSSLW 165
Score = 25.8 bits (55), Expect(2) = 2e-05
Identities = 17/44 (38%), Positives = 17/44 (38%)
Frame = +3
Query: 244 MFGFQKHNVANQREHLILLLANVHIRQFPNPDQQPKLDECALTE 287
MFGFQK NV N P DQ K DE L E
Sbjct: 15 MFGFQKDNVEN-----------------PKSDQLLKFDESCLNE 95
>TC218628 similar to UP|Q9LWU7 (Q9LWU7) ESTs AU033035(S1515), partial (13%)
Length = 1115
Score = 44.3 bits (103), Expect = 5e-04
Identities = 45/125 (36%), Positives = 62/125 (49%)
Frame = +2
Query: 1690 LSRVLSSWYYLLYTKFLVIHTEVLLHIY*LLYQCGSWWEPGCLLHSCSTPLGLNGKKLLM 1749
LS+ L +L + + + EVLL ++ L GS G L + STPL +GK+L
Sbjct: 2 LSKRLRWHCFL*FISHMGMQREVLLRMFC*LLAVGSLSFLGFSLLTFSTPLVSSGKRLWK 181
Query: 1750 IGLIGINGLVTEEV*VYRLKKVGNLGGKKNKIIYSIQESGES*WRYCCHCDFLSTNMVLY 1809
I +IG G T+E * + VG+LGG KN+ I + G WR NMVL
Sbjct: 182 ILMIGQVGFFTKEE*E*KEIIVGSLGGTKNRCISKL--*GVVFWRQF*VQGSSCFNMVLC 355
Query: 1810 IT*IL 1814
I+ IL
Sbjct: 356 ISCIL 370
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.348 0.154 0.546
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 103,822,887
Number of Sequences: 63676
Number of extensions: 1733851
Number of successful extensions: 18186
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 13016
Number of HSP's successfully gapped in prelim test: 491
Number of HSP's that attempted gapping in prelim test: 4210
Number of HSP's gapped (non-prelim): 14843
length of query: 1982
length of database: 12,639,632
effective HSP length: 111
effective length of query: 1871
effective length of database: 5,571,596
effective search space: 10424456116
effective search space used: 10424456116
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC122723.7


