

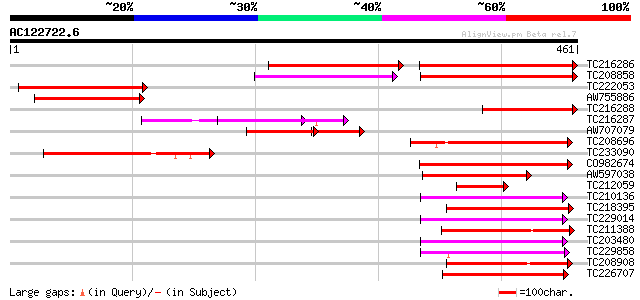
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122722.6 + phase: 0
(461 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216286 similar to UP|Q8GTR0 (Q8GTR0) Sugar transporter, partia... 217 1e-97
TC208858 similar to UP|Q9M9Z8 (Q9M9Z8) F20B17.24, partial (51%) 177 3e-54
TC222053 similar to UP|Q8GTR0 (Q8GTR0) Sugar transporter, partia... 185 3e-47
AW755886 similar to GP|27261731|gb| sugar transporter {Citrus un... 146 2e-35
TC216288 homologue to UP|Q8GTR0 (Q8GTR0) Sugar transporter, part... 143 2e-34
TC216287 similar to UP|Q8GTR0 (Q8GTR0) Sugar transporter, partia... 138 6e-33
AW707079 similar to GP|27261731|gb| sugar transporter {Citrus un... 94 1e-32
TC208696 similar to UP|Q93WT7 (Q93WT7) Hexose transporter pGlT, ... 119 3e-27
TC233090 similar to UP|Q9ZVN7 (Q9ZVN7) T7A14.10 protein, partial... 115 5e-26
CO982674 107 9e-24
AW597038 95 7e-20
TC212059 similar to UP|Q9FYG3 (Q9FYG3) F1N21.12, partial (10%) 79 4e-15
TC210136 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, par... 77 1e-14
TC218395 similar to UP|Q9FIF2 (Q9FIF2) Sugar transporter-like pr... 71 9e-13
TC229014 similar to UP|Q8GT52 (Q8GT52) Hexose transporter, parti... 70 2e-12
TC211388 similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-like pr... 70 2e-12
TC203480 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, par... 69 3e-12
TC229858 weakly similar to UP|Q8VHD6 (Q8VHD6) Glucose transporte... 68 1e-11
TC208908 weakly similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-... 66 4e-11
TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter... 65 6e-11
>TC216286 similar to UP|Q8GTR0 (Q8GTR0) Sugar transporter, partial (45%)
Length = 1342
Score = 217 bits (553), Expect(2) = 1e-97
Identities = 111/128 (86%), Positives = 117/128 (90%)
Frame = +2
Query: 334 AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAM 393
AI+MI+QATGA++L+ GA Y SVGGM LFV TFALGAGPVPGLLL EIFPSRIRAKAM
Sbjct: 335 AIAMILQATGATSLVSNMGAQYFSVGGMFLFVLTFALGAGPVPGLLLPEIFPSRIRAKAM 514
Query: 394 AFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
A CMSVHWVINFFVGLLFLRLLEKLG QLLYSMFATFCIMAVIFVKRNVVETKGKSL EI
Sbjct: 515 AVCMSVHWVINFFVGLLFLRLLEKLGPQLLYSMFATFCIMAVIFVKRNVVETKGKSLHEI 694
Query: 454 EIALLPQE 461
EIALLPQ+
Sbjct: 695 EIALLPQD 718
Score = 157 bits (398), Expect(2) = 1e-97
Identities = 84/110 (76%), Positives = 91/110 (82%)
Frame = +3
Query: 211 FWVYPKQNLRCHSYPR*IEEKILTL*SSLNCFMAIILKLFLLDQPYLLYNSYLV*TLCFI 270
F VY KQNL+C SYPR IEE I+ +*S NCFM +ILKLFLLDQPYLLYNSYLV* LCFI
Sbjct: 3 FLVYQKQNLQCQSYPRQIEETIVIV*SCRNCFMVVILKLFLLDQPYLLYNSYLV*MLCFI 182
Query: 271 SLQLFLKVPECHRILQTSA*ELRI*QDLSFRRV*WINLEGRFYSFGVSLA 320
SL LFLKV ECH+ LQ SA*EL I QDLSF+ V*WI+LEGRFY GVSLA
Sbjct: 183 SLLLFLKVLECHQTLQMSA*ELPIWQDLSFQWV*WISLEGRFYFSGVSLA 332
>TC208858 similar to UP|Q9M9Z8 (Q9M9Z8) F20B17.24, partial (51%)
Length = 993
Score = 177 bits (449), Expect(2) = 3e-54
Identities = 87/127 (68%), Positives = 106/127 (82%)
Frame = +3
Query: 335 ISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMA 394
+SM +Q AS+ G++YLSVGGMLLFV +FA GAGPVP L+++EI PS IRAKAMA
Sbjct: 369 LSMGLQVIAASSFASGFGSMYLSVGGMLLFVLSFAFGAGPVPCLIMSEILPSNIRAKAMA 548
Query: 395 FCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIE 454
C++VHWVINFFVGL FLRLLE +GAQLLYS+F C++AV+FVK+N++ETKGKSLQEIE
Sbjct: 549 ICLAVHWVINFFVGLFFLRLLELIGAQLLYSIFGFCCLIAVVFVKKNILETKGKSLQEIE 728
Query: 455 IALLPQE 461
IALL QE
Sbjct: 729 IALLAQE 749
Score = 53.1 bits (126), Expect(2) = 3e-54
Identities = 48/116 (41%), Positives = 59/116 (50%)
Frame = +1
Query: 200 ELLKQKLSLKDFWVYPKQNLRCHSYPR*IEEKILTL*SSLNCFMAIILKLFLLDQPYLLY 259
E LK KL L+ W NL+ + E L *+ N A I + LD +L
Sbjct: 1 EPLKLKLHLRSSWGEYM*NLQ*LNCQSLTEVMDLIQ*NFQN*STAAISE*CSLDLHFLRC 180
Query: 260 NSYLV*TLCFISLQLFLKVPECHRILQTSA*ELRI*QDLSFRRV*WINLEGRFYSF 315
+SYL * L ISLQL LKV H ILQT *E I L + *WINLEG+ +S+
Sbjct: 181 SSYLA*MLFSISLQLSLKVLVYHLILQTHV*EFAIYWGL*LQ*F*WINLEGKCFSW 348
>TC222053 similar to UP|Q8GTR0 (Q8GTR0) Sugar transporter, partial (19%)
Length = 484
Score = 185 bits (470), Expect = 3e-47
Identities = 93/105 (88%), Positives = 97/105 (91%)
Frame = +3
Query: 8 DLLDNFIDKETTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAE 67
DLLD +DK T+NPS LSLPHVLVATI+SFLFGYHLGVVNEPLESISVDLGF GNTLAE
Sbjct: 165 DLLDIGLDKGTSNPSLMLSLPHVLVATISSFLFGYHLGVVNEPLESISVDLGFRGNTLAE 344
Query: 68 GLVVSICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAMS 112
GLVVSICLGGAL GCLLSGWIAD VGRRRAFQLCALPMIIGA+MS
Sbjct: 345 GLVVSICLGGALIGCLLSGWIADGVGRRRAFQLCALPMIIGASMS 479
>AW755886 similar to GP|27261731|gb| sugar transporter {Citrus unshiu},
partial (18%)
Length = 437
Score = 146 bits (368), Expect = 2e-35
Identities = 68/89 (76%), Positives = 79/89 (88%)
Frame = +3
Query: 21 PSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALF 80
P W+ SL HV+VA+++SFL+GYH+GVVNE LESIS+DLGF+GNT+AEGLVVSICLGGA
Sbjct: 171 PCWRRSLRHVIVASLSSFLYGYHIGVVNETLESISIDLGFSGNTMAEGLVVSICLGGAFV 350
Query: 81 GCLLSGWIADAVGRRRAFQLCALPMIIGA 109
G L SGWIAD VGR R+FQLCALPMIIGA
Sbjct: 351 GSLFSGWIADGVGRXRSFQLCALPMIIGA 437
>TC216288 homologue to UP|Q8GTR0 (Q8GTR0) Sugar transporter, partial (16%)
Length = 886
Score = 143 bits (360), Expect = 2e-34
Identities = 72/77 (93%), Positives = 73/77 (94%)
Frame = +1
Query: 385 PSRIRAKAMAFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVE 444
PSRIRAKAMA CMSVHWVINFFVGLLFLRLLEKLG QLLYSMFATFCIMAV FVKRNVVE
Sbjct: 1 PSRIRAKAMAVCMSVHWVINFFVGLLFLRLLEKLGPQLLYSMFATFCIMAVTFVKRNVVE 180
Query: 445 TKGKSLQEIEIALLPQE 461
TKGKSL EIEIALLPQ+
Sbjct: 181 TKGKSLHEIEIALLPQD 231
>TC216287 similar to UP|Q8GTR0 (Q8GTR0) Sugar transporter, partial (37%)
Length = 696
Score = 138 bits (347), Expect = 6e-33
Identities = 81/149 (54%), Positives = 88/149 (58%), Gaps = 43/149 (28%)
Frame = +1
Query: 170 ISGCLPYLQLYLLWLWSSVQRVHIGYTSKEELLKQKLSLKDFWVYPKQNLRCHSYPR*IE 229
+ G LP+ LYL LWSS QRVHIGYTSKEELLKQKLSL+ F VY KQNL+C SYPR IE
Sbjct: 250 VFGYLPFQLLYLPQLWSSAQRVHIGYTSKEELLKQKLSLRGFLVYQKQNLQCQSYPRQIE 429
Query: 230 EKILTL*SSLNCFMAIILK----------------------------------------- 248
E I+ +*S NCFM +ILK
Sbjct: 430 ETIVIV*SCRNCFMVVILKVCIFPGLSLG*L*LVSVFAIVVIWSLVSLGFQKLFFIHKFV 609
Query: 249 --LFLLDQPYLLYNSYLV*TLCFISLQLF 275
LFLLDQPYLLYNSYLV* LCFISLQLF
Sbjct: 610 LQLFLLDQPYLLYNSYLV*MLCFISLQLF 696
Score = 67.8 bits (164), Expect = 1e-11
Identities = 45/134 (33%), Positives = 58/134 (42%)
Frame = +2
Query: 108 GAAMSNKQLVWHACRKIICWNWLGLGPSRCCSLCDRAYGALIQIATCFGILGSLFIGIPV 167
G NKQLVWHACRK++CW+WLG GPS C S+CDR + C G L S +
Sbjct: 17 GYECGNKQLVWHACRKVVCWDWLGPGPSCCLSVCDRGFSCF-----CAGHLWSFHPDCHM 181
Query: 168 KEISGCLPYLQLYLLWLWSSVQRVHIGYTSKEELLKQKLSLKDFWVYPKQNLRCHSYPR* 227
+G Y W+ QR +I + L +Y CHSY
Sbjct: 182 PWSNGSFIY--------WNPCQR-NIWMVAGLFL----------GIYHSSCYTCHSYGLL 304
Query: 228 IEEKILTL*SSLNC 241
E L + + NC
Sbjct: 305 RRESTLVIQARKNC 346
>AW707079 similar to GP|27261731|gb| sugar transporter {Citrus unshiu},
partial (17%)
Length = 419
Score = 94.0 bits (232), Expect(2) = 1e-32
Identities = 45/59 (76%), Positives = 51/59 (86%)
Frame = +1
Query: 193 IGYTSKEELLKQKLSLKDFWVYPKQNLRCHSYPR*IEEKILTL*SSLNCFMAIILKLFL 251
IGYTSKEELLKQKLSL+DFWVY KQNL+C SYPR*IE IL L*S NCFM +ILK+++
Sbjct: 10 IGYTSKEELLKQKLSLRDFWVYQKQNLQCRSYPR*IEVMILIL*SCQNCFMVVILKVYI 186
Score = 64.3 bits (155), Expect(2) = 1e-32
Identities = 33/43 (76%), Positives = 37/43 (85%)
Frame = +3
Query: 246 ILKLFLLDQPYLLYNSYLV*TLCFISLQLFLKVPECHRILQTS 288
IL+LFLL+QPYLLYNSYL+* L FISLQLFLKV E H+ LQ S
Sbjct: 291 ILQLFLLNQPYLLYNSYLI*MLYFISLQLFLKVLESHQTLQIS 419
>TC208696 similar to UP|Q93WT7 (Q93WT7) Hexose transporter pGlT, partial
(23%)
Length = 1373
Score = 119 bits (298), Expect = 3e-27
Identities = 62/133 (46%), Positives = 89/133 (66%), Gaps = 2/133 (1%)
Frame = +1
Query: 327 YIFCTV*AISMIIQATGAS--TLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIF 384
+ C A SM++ + + L P +G L +V G +L+V +F+LGAGPVP LLL EIF
Sbjct: 625 FSLCLTQAASMLLLSLSFTWKVLAPYSGTL--AVLGTVLYVLSFSLGAGPVPALLLPEIF 798
Query: 385 PSRIRAKAMAFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVE 444
SRIRAKA++ + HW+ NF +GL FL ++ K G +Y F+ C++AV+++ NVVE
Sbjct: 799 ASRIRAKAVSLSLGTHWISNFVIGLYFLSVVNKFGISSVYLGFSAVCVLAVLYIAGNVVE 978
Query: 445 TKGKSLQEIEIAL 457
TKG+SL+EIE AL
Sbjct: 979 TKGRSLEEIERAL 1017
>TC233090 similar to UP|Q9ZVN7 (Q9ZVN7) T7A14.10 protein, partial (22%)
Length = 594
Score = 115 bits (287), Expect = 5e-26
Identities = 66/152 (43%), Positives = 95/152 (62%), Gaps = 13/152 (8%)
Frame = +3
Query: 28 PHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSGW 87
PHVLVA++++F+FGYH+GV+N P+ SI+ +LGF GN+ EGLVVSI + GA G + S
Sbjct: 6 PHVLVASMSNFIFGYHIGVMNGPIVSIARELGFEGNSFIEGLVVSIFIAGAFIGSISSAS 185
Query: 88 IADAVGRRRAFQLCALPMIIGAAMSNKQLVWHACRKIICWNWL-GLG--------PSRCC 138
+ D +G R FQ+ ++P+I+GA +S + H+ +II +L GLG P
Sbjct: 186 LLDRLGSRLTFQINSIPLILGAIISAQA---HSLNEIIGGRFLVGLGIGVNTVLVPIYIS 356
Query: 139 SLCDRAY----GALIQIATCFGILGSLFIGIP 166
+ Y G+L QI TC GI+ SLF+GIP
Sbjct: 357 EVAPTKYRGALGSLCQIGTCLGIITSLFLGIP 452
>CO982674
Length = 703
Score = 107 bits (268), Expect = 9e-24
Identities = 54/124 (43%), Positives = 78/124 (62%)
Frame = -1
Query: 334 AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAM 393
AISM + A+G L LS+ G ++++F+FA+GAGPV G+++ E+ +R R K M
Sbjct: 514 AISMFLVASGIIFPLDEQLGNNLSILGTIMYIFSFAIGAGPVTGIIIPELSSTRTRGKIM 335
Query: 394 AFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
F S HWV NF VGL FL L++K G +Y+ F ++A F +VETKG+SL+EI
Sbjct: 334 GFSFSTHWVCNFVVGLFFLELVDKFGVAPVYASFGAISLLAATFAYYFIVETKGRSLEEI 155
Query: 454 EIAL 457
E +L
Sbjct: 154 ERSL 143
>AW597038
Length = 283
Score = 94.7 bits (234), Expect = 7e-20
Identities = 48/89 (53%), Positives = 62/89 (68%)
Frame = +2
Query: 336 SMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAF 395
SM +Q A + G++YLS GGMLLF +FA AGPV L+++EI PS IR+KAMA
Sbjct: 17 SMGVQVIAACSFAS*LGSMYLSAGGMLLFELSFAFSAGPVTCLIMSEILPSNIRSKAMAI 196
Query: 396 CMSVHWVINFFVGLLFLRLLEKLGAQLLY 424
C++VH VINFF GL ++ LLE +GA LY
Sbjct: 197 CLAVH*VINFFWGLSYMLLLELIGAHTLY 283
>TC212059 similar to UP|Q9FYG3 (Q9FYG3) F1N21.12, partial (10%)
Length = 423
Score = 79.0 bits (193), Expect = 4e-15
Identities = 38/42 (90%), Positives = 38/42 (90%)
Frame = +3
Query: 364 FVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINF 405
FV TFALGAGPVPGLLL EIFPSRIRAKAMA CMSVHWVI F
Sbjct: 246 FVLTFALGAGPVPGLLLPEIFPSRIRAKAMAVCMSVHWVIFF 371
>TC210136 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, partial (19%)
Length = 623
Score = 77.4 bits (189), Expect = 1e-14
Identities = 37/119 (31%), Positives = 65/119 (54%)
Frame = +2
Query: 335 ISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMA 394
+S++I G+ L + ++S ++++ F +G GP+P +L +EIFP+R+R +A
Sbjct: 41 VSLLILVIGSLVELDSTINAFISTSSVIVYFCCFVMGFGPIPNILCSEIFPTRVRGLCIA 220
Query: 395 FCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
C W+ + V +L +G ++ M+A CI+A +FV V ETKG L+ I
Sbjct: 221 ICALTFWICDIIVTYTLPVMLNSVGLGGVFGMYAVVCIIAWVFVFLKVPETKGMPLEVI 397
>TC218395 similar to UP|Q9FIF2 (Q9FIF2) Sugar transporter-like protein,
partial (40%)
Length = 1013
Score = 71.2 bits (173), Expect = 9e-13
Identities = 35/103 (33%), Positives = 63/103 (60%)
Frame = +1
Query: 356 LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLL 415
++VG +LL+V + + GP+ L+++E+FP R R K ++ + ++ N V F L
Sbjct: 409 VAVGALLLYVGCYQISFGPISWLMVSEVFPLRTRGKGISLAVLTNFASNAVVTFAFSPLK 588
Query: 416 EKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIALL 458
E LGA+ L+ +F ++++F+ +V ETKG SL++IE +L
Sbjct: 589 EFLGAENLFLLFGAIATLSLLFIIFSVPETKGMSLEDIESKIL 717
>TC229014 similar to UP|Q8GT52 (Q8GT52) Hexose transporter, partial (23%)
Length = 714
Score = 70.5 bits (171), Expect = 2e-12
Identities = 36/119 (30%), Positives = 60/119 (50%)
Frame = +2
Query: 335 ISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMA 394
++++I G+ L T +S + + F +G GP+P +L EIFP+R+R +A
Sbjct: 122 VALLILVLGSLVDLGTTANASISTISVXXYFCFFVMGFGPIPNILCAEIFPTRVRGLCIA 301
Query: 395 FCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
C W+ + V +L LG ++ ++A C +A +FV V ETKG L+ I
Sbjct: 302 ICALTFWICDIIVTYTLPVMLNSLGLAGVFGIYAVACFIAWVFVFLKVPETKGMPLEVI 478
>TC211388 similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-like protein,
partial (27%)
Length = 786
Score = 70.1 bits (170), Expect = 2e-12
Identities = 33/108 (30%), Positives = 67/108 (61%)
Frame = +3
Query: 352 GALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLF 411
G+ L++ G+L++ +F+LG G +P ++++EIFP+ ++ A + V W+ ++ V F
Sbjct: 231 GSPILALAGVLVYTGSFSLGMGGIPWVIMSEIFPTNVKGSAGSLVTLVSWLCSWIVSYAF 410
Query: 412 LRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIALLP 459
L+ A + +F++ C ++FV + V ETKG++L+E++ +L P
Sbjct: 411 NFLMSWSSAGTFF-IFSSICGFTILFVAKLVPETKGRTLEEVQASLNP 551
>TC203480 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, partial (38%)
Length = 1040
Score = 69.3 bits (168), Expect = 3e-12
Identities = 36/119 (30%), Positives = 63/119 (52%)
Frame = +2
Query: 335 ISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMA 394
+S+II G+ +S ++++ F +G GP+P +L +EIFP+R+R +A
Sbjct: 467 VSLIILVIGSLVNFGNVAHAAISTVCVVVYFCCFVMGYGPIPNILCSEIFPTRVRGLCIA 646
Query: 395 FCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
C V W+ + + +L LG +++++A C ++ IFV V ETKG L+ I
Sbjct: 647 ICALVFWIGDIIITYSLPVMLGSLGLGGVFAIYAVVCFISWIFVFLKVPETKGMPLEVI 823
>TC229858 weakly similar to UP|Q8VHD6 (Q8VHD6) Glucose transporter 10 (Mus
musculus 13 days embryo heart cDNA, RIKEN full-length
enriched library, clone:D330001H08 product:solute
carrier family 2 (facilitated glucose transporter),
member 10, full insert sequence), partial (4%)
Length = 715
Score = 67.8 bits (164), Expect = 1e-11
Identities = 40/123 (32%), Positives = 62/123 (49%), Gaps = 2/123 (1%)
Frame = +3
Query: 335 ISMIIQATGASTLLPTAGALY--LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKA 392
+S++I A T+ LY L+V G+ L++ F+ G GPVP L +EI+P R
Sbjct: 249 VSLVILAFAFYKQSSTSNELYGWLAVVGLALYIGFFSPGMGPVPWTLSSEIYPEEYRGIC 428
Query: 393 MAFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQE 452
+V WV N V FL + E +G + + ++A +FV V ETKG + E
Sbjct: 429 GGMSATVCWVSNLIVSETFLSIAEGIGIGSTFLIIGVIAVVAFVFVLVYVPETKGLTFDE 608
Query: 453 IEI 455
+E+
Sbjct: 609 VEV 617
>TC208908 weakly similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-like
protein, partial (28%)
Length = 753
Score = 65.9 bits (159), Expect = 4e-11
Identities = 32/102 (31%), Positives = 64/102 (62%)
Frame = +2
Query: 356 LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLL 415
L++ G+L++V ++++G G +P ++++EIFP ++ A + V W+ ++ + F L+
Sbjct: 182 LALVGVLVYVGSYSIGMGAIPWVIMSEIFPINVKGSAGSLVTLVSWLCSWIISYSFNFLM 361
Query: 416 EKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIAL 457
A + MF++ C V+FV + V ETKG++L+EI+ +L
Sbjct: 362 SWSSAG-TFLMFSSICGFTVLFVAKLVPETKGRTLEEIQASL 484
>TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 2338
Score = 65.1 bits (157), Expect = 6e-11
Identities = 36/102 (35%), Positives = 63/102 (61%)
Frame = +3
Query: 353 ALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFL 412
A+ LS+ +L +V TF++G+GP+ + +EIFP R+RA+ +A +V+ V + + + FL
Sbjct: 657 AVGLSIAAVLSYVATFSIGSGPITWVYSSEIFPLRLRAQGVAIGAAVNRVTSGVIAMTFL 836
Query: 413 RLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIE 454
L + + + +FA +A IF + ET+GK+L+EIE
Sbjct: 837 SLQKAITIGGAFFLFAGVAAVAWIFHYTLLPETRGKTLEEIE 962
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.341 0.151 0.500
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,100,575
Number of Sequences: 63676
Number of extensions: 419390
Number of successful extensions: 3793
Number of sequences better than 10.0: 90
Number of HSP's better than 10.0 without gapping: 3745
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3784
length of query: 461
length of database: 12,639,632
effective HSP length: 101
effective length of query: 360
effective length of database: 6,208,356
effective search space: 2235008160
effective search space used: 2235008160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122722.6


