

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
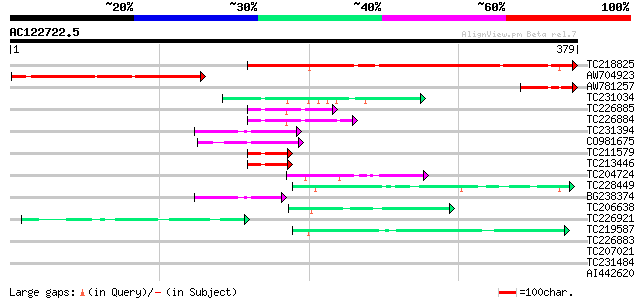
Query= AC122722.5 - phase: 0
(379 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC218825 weakly similar to GB|AAD09747.1|4191329|AF065392 ZIS2 {... 344 4e-95
AW704923 164 6e-41
AW781257 55 4e-08
TC231034 similar to UP|Q8S9M8 (Q8S9M8) P53 binding protein (Frag... 52 4e-07
TC226885 similar to GB|AAO63346.1|28950845|BT005282 At1g67325 {A... 48 8e-06
TC226884 similar to GB|AAO63346.1|28950845|BT005282 At1g67325 {A... 48 8e-06
TC231394 similar to UP|Q9LW11 (Q9LW11) Zinc finger protein-like;... 47 1e-05
CO981675 46 3e-05
TC211579 similar to UP|Q8S8K1 (Q8S8K1) Predicted protein, partia... 44 9e-05
TC213446 similar to UP|Q8S8K1 (Q8S8K1) Predicted protein, partia... 44 9e-05
TC204724 similar to UP|Q9AXQ2 (Q9AXQ2) Mitochondrial processing ... 44 1e-04
TC228449 similar to GB|AAR99134.1|41058179|BT011476 RE11923p {Dr... 44 2e-04
BG238374 similar to GP|11994340|db zinc finger protein-like; Ser... 44 2e-04
TC206638 similar to UP|Q9FYA8 (Q9FYA8) SC35-like splicing factor... 42 5e-04
TC226921 similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein p... 42 5e-04
TC219587 similar to PIR|T49181|T49181 cyclophylin-like protein -... 41 8e-04
TC226883 similar to GB|AAO63346.1|28950845|BT005282 At1g67325 {A... 40 0.001
TC207021 similar to UP|Q940Z6 (Q940Z6) At1g12810/F13K23_4, parti... 40 0.002
TC231484 similar to UP|Q6ZLD4 (Q6ZLD4) Cyclophylin-like protein,... 40 0.002
AI442620 40 0.002
>TC218825 weakly similar to GB|AAD09747.1|4191329|AF065392 ZIS2 {Homo
sapiens;} , partial (8%)
Length = 833
Score = 344 bits (882), Expect = 4e-95
Identities = 168/224 (75%), Positives = 183/224 (81%), Gaps = 4/224 (1%)
Frame = +1
Query: 160 REGDWMCPDALCGNLNFARRDFCNQCKRPRPAAAGSPPRR--GSPPLHAPPRRYPGPPFD 217
REGDW+CPD LC NLNFARRD CN C R R A A SP R PPLHAPPRR+PGP D
Sbjct: 4 REGDWICPDPLCNNLNFARRDHCNSCNRSRNAPARSPRRAYPAPPPLHAPPRRFPGPLID 183
Query: 218 RSPERPMNGYRSPPPRLMGRDGLRDYGPAAAALPPLRHEGRFPDPHLHRERMDYMDDAYR 277
RSPER +NGYRSP R + RDG R+YG +AALPPLRHEGRFPDPH+ RERMD+++DAYR
Sbjct: 184 RSPERTLNGYRSP--RGLARDGPREYG--SAALPPLRHEGRFPDPHVRRERMDFIEDAYR 351
Query: 278 GRGKFDRPPPLDWDNRDRGRDGFSNERKGFERRPLSPSAPLLPSLPPHRGGDRWSRDVRD 337
GR KFDRPPPLDWDNRDRGRDGFS+ERKGFERRPLSP P LPSLP HRGG RW+RDVR+
Sbjct: 352 GRNKFDRPPPLDWDNRDRGRDGFSDERKGFERRPLSPPVPFLPSLPHHRGG-RWARDVRE 528
Query: 338 RSRSPIRGGPPAKDYRRDPVMSRGGRDDR--RGGVGRDRIGGMY 379
RSRSPIRGGPP K+YRRD M R RDDR R G+GRDRIGGMY
Sbjct: 529 RSRSPIRGGPPPKEYRRDTFMHR-ARDDRHDRHGMGRDRIGGMY 657
>AW704923
Length = 404
Score = 164 bits (415), Expect = 6e-41
Identities = 83/130 (63%), Positives = 93/130 (70%)
Frame = +1
Query: 2 SSTSPSQSPPSHHHHHQPLLSSLVVRPSLTENSPPGAAAHSNDYEPGELRRDPPPVNYSR 61
SS +PP HHH PLLSSLVVRPS T+ + + D+EPGEL RDPPP YSR
Sbjct: 28 SSRDKEPAPP---HHHPPLLSSLVVRPSHTDAAAAAGGGRAADFEPGELHRDPPPP-YSR 195
Query: 62 SDRYSDDAGYRFRAGSSSPVHHRRDADHRFPSDYNHFPRNRGFGGGRDFGRFRDPPHPYA 121
SDRY D+ GYR RAGSSSPVH RRDAD RF SDYN+ R+RG+GGGRD GR+RDP PY
Sbjct: 196 SDRYPDEPGYRIRAGSSSPVH-RRDADRRFASDYNNLSRSRGYGGGRDPGRYRDPSPPYG 372
Query: 122 RGRPGGRPFG 131
RGR GG P G
Sbjct: 373 RGRVGGMPMG 402
>AW781257
Length = 265
Score = 55.5 bits (132), Expect = 4e-08
Identities = 28/38 (73%), Positives = 30/38 (78%)
Frame = +3
Query: 342 PIRGGPPAKDYRRDPVMSRGGRDDRRGGVGRDRIGGMY 379
PIRGGPP K+YRRD M R DDRR G+GRDRIGGMY
Sbjct: 6 PIRGGPPPKEYRRDTFMHR-ATDDRR-GMGRDRIGGMY 113
>TC231034 similar to UP|Q8S9M8 (Q8S9M8) P53 binding protein (Fragment),
partial (71%)
Length = 892
Score = 52.0 bits (123), Expect = 4e-07
Identities = 51/174 (29%), Positives = 67/174 (38%), Gaps = 38/174 (21%)
Frame = +3
Query: 143 HGRGEGLHGRNNPNVRPREGDWMCPDALCGNLNFARRDFCNQ--CKRPRPAAAGSPPR-- 198
+ RG R + ++GDW CP+ CGNLNF+ R CNQ C PRP+ +PP
Sbjct: 57 NNRGSPGSKRFRNDASRKDGDWTCPN--CGNLNFSFRTVCNQGHCGAPRPSLTLTPPAPI 230
Query: 199 ----RGSPPLH-----APPRRY------------PGPPFD-------RSPERPMNGYRSP 230
R S P + PP Y PG +D R+P P+ +
Sbjct: 231 ISPYRNSHPFYHGGVGIPPPSYGIPSQFGSPIPHPGIQYDYGLYARARAPYSPLPMF--- 401
Query: 231 PPRLMG------RDGLRDYGPAAAALPPLRHEGRFPDPHLHRERMDYMDDAYRG 278
PP G R + YG + PP EG D R+R D G
Sbjct: 402 PPASFGGINYGPRPRIDGYGYGFQSPPPPWAEGLVADNFASRKRRGGPDGLSEG 563
Score = 34.7 bits (78), Expect = 0.073
Identities = 34/125 (27%), Positives = 44/125 (35%), Gaps = 25/125 (20%)
Frame = +3
Query: 94 DYNHFPRNRGFGGGRDFGRFRDPPHPYARGRPGGRPFGRAFDGPGYGHRHGR-------- 145
+Y PR G+G G F+ PP P+A G R G G G
Sbjct: 426 NYGPRPRIDGYGYG-----FQSPPPPWAEGLVADNFASRKRRGGPDGLSEGDWICPRCDN 590
Query: 146 --------------GEGLHGRNNPNVRP-REGDWMCPDALCGNLNFARRDFCNQ--CKRP 188
G N PN EG W C CGNLN+ R+ CN+ C+
Sbjct: 591 VNFAFRTTCNIKHCGAVKPSSNKPNTAVIPEGSWTCEK--CGNLNYPFRNVCNRKDCRNE 764
Query: 189 RPAAA 193
+ +A
Sbjct: 765 KTVSA 779
>TC226885 similar to GB|AAO63346.1|28950845|BT005282 At1g67325 {Arabidopsis
thaliana;} , partial (27%)
Length = 388
Score = 47.8 bits (112), Expect = 8e-06
Identities = 27/62 (43%), Positives = 33/62 (52%), Gaps = 2/62 (3%)
Frame = +2
Query: 160 REGDWMCPDALCGNLNFARRDFCN--QCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFD 217
RE DW CP CGN+NF+ R CN C +PRPA S + + PL AP P+
Sbjct: 152 REDDWTCPS--CGNVNFSFRTTCNMRNCTQPRPADHNS--KSAAKPLQAPQGYSSSAPYL 319
Query: 218 RS 219
RS
Sbjct: 320 RS 325
>TC226884 similar to GB|AAO63346.1|28950845|BT005282 At1g67325 {Arabidopsis
thaliana;} , partial (49%)
Length = 503
Score = 47.8 bits (112), Expect = 8e-06
Identities = 30/75 (40%), Positives = 37/75 (49%), Gaps = 2/75 (2%)
Frame = +3
Query: 160 REGDWMCPDALCGNLNFARRDFCN--QCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFD 217
RE DW CP CGN+NF+ R CN C +PRPA S + + PL AP P+
Sbjct: 144 REDDWTCPS--CGNVNFSFRTTCNMRNCTQPRPADHNS--KSAAKPLQAPQGYSSSAPYL 311
Query: 218 RSPERPMNGYRSPPP 232
S P + Y PP
Sbjct: 312 GS-NTPSSIYLGVPP 353
>TC231394 similar to UP|Q9LW11 (Q9LW11) Zinc finger protein-like; Ser/Thr
protein kinase-like protein, partial (62%)
Length = 689
Score = 47.0 bits (110), Expect = 1e-05
Identities = 30/72 (41%), Positives = 35/72 (47%)
Frame = +2
Query: 124 RPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLNFARRDFCN 183
R G +G D G+G R G GL G + VRP GDW C A CG NFA R C
Sbjct: 44 RCGDAKYGDRVDFGGFGGRGGSSFGLTGSD---VRP--GDWYCAAANCGAHNFASRSSCF 208
Query: 184 QCKRPRPAAAGS 195
+C + AGS
Sbjct: 209 KCGAFKDDLAGS 244
Score = 33.9 bits (76), Expect = 0.12
Identities = 13/30 (43%), Positives = 17/30 (56%)
Frame = +2
Query: 160 REGDWMCPDALCGNLNFARRDFCNQCKRPR 189
+ GDW+C + C NFA R C +C PR
Sbjct: 305 KSGDWICTRSGCNEHNFASRMECFKCSAPR 394
>CO981675
Length = 810
Score = 45.8 bits (107), Expect = 3e-05
Identities = 27/72 (37%), Positives = 33/72 (45%), Gaps = 1/72 (1%)
Frame = -3
Query: 126 GGRPFGRAFDGPGYGHRHGRGEGLHG-RNNPNVRPREGDWMCPDALCGNLNFARRDFCNQ 184
GG +G F G GRG G P+VRP GDW C CG NFA R C +
Sbjct: 661 GGGDYGGGFGG-------GRGSSSFGFTTGPDVRP--GDWYCTVGNCGAHNFASRSSCFK 509
Query: 185 CKRPRPAAAGSP 196
C P+ ++ P
Sbjct: 508 CGAPKEDSSAGP 473
Score = 38.1 bits (87), Expect = 0.007
Identities = 17/53 (32%), Positives = 26/53 (48%)
Frame = -3
Query: 145 RGEGLHGRNNPNVRPREGDWMCPDALCGNLNFARRDFCNQCKRPRPAAAGSPP 197
R G G ++ + GDW+C + C NFA R C +C PR +++ P
Sbjct: 448 RPYGFGGGSSARPGWKSGDWICTRSGCNEHNFANRMECYRCNAPRDSSSARSP 290
>TC211579 similar to UP|Q8S8K1 (Q8S8K1) Predicted protein, partial (20%)
Length = 742
Score = 44.3 bits (103), Expect = 9e-05
Identities = 18/30 (60%), Positives = 21/30 (70%)
Frame = +3
Query: 160 REGDWMCPDALCGNLNFARRDFCNQCKRPR 189
REGDW C C N N+A R FCN+CK+PR
Sbjct: 180 REGDWECSS--CNNRNYAFRSFCNRCKQPR 263
>TC213446 similar to UP|Q8S8K1 (Q8S8K1) Predicted protein, partial (37%)
Length = 424
Score = 44.3 bits (103), Expect = 9e-05
Identities = 18/30 (60%), Positives = 21/30 (70%)
Frame = +3
Query: 160 REGDWMCPDALCGNLNFARRDFCNQCKRPR 189
REGDW C C N N+A R FCN+CK+PR
Sbjct: 51 REGDWECSS--CNNRNYAFRSFCNRCKQPR 134
Score = 38.9 bits (89), Expect = 0.004
Identities = 23/67 (34%), Positives = 31/67 (45%)
Frame = +3
Query: 159 PREGDWMCPDALCGNLNFARRDFCNQCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFDR 218
PR GDW+C C N N+A R+ C +C +P+ AA P PP +
Sbjct: 177 PRIGDWICTG--CTNNNYASREKCKKCGQPKEVAA--MPAIAMTGASFPPYSHYFSRAPG 344
Query: 219 SPERPMN 225
PE+ MN
Sbjct: 345 GPEQKMN 365
>TC204724 similar to UP|Q9AXQ2 (Q9AXQ2) Mitochondrial processing peptidase
beta subunit, partial (92%)
Length = 1868
Score = 43.9 bits (102), Expect = 1e-04
Identities = 36/104 (34%), Positives = 47/104 (44%), Gaps = 9/104 (8%)
Frame = +2
Query: 186 KRPRPAAAGSP------PRRGSPPLHAPPRRYPGPPFDRS---PERPMNGYRSPPPRLMG 236
+R P+ SP PRR + AP R P PP ++ P P+ + P L+
Sbjct: 158 RRLLPSRPSSPHRHDLRPRRRGREVQAPAAREPRPPLPQARVPPAHPL*PHAHP---LLP 328
Query: 237 RDGLRDYGPAAAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRG 280
RD RD+ A+ PP RH PH HR R+D A R RG
Sbjct: 329 RDA-RDH---ASQRPPHRHRVHPQRPHRHRRRLDRRRFAVRDRG 448
Score = 29.3 bits (64), Expect = 3.1
Identities = 30/92 (32%), Positives = 34/92 (36%), Gaps = 19/92 (20%)
Frame = +1
Query: 188 PRPAA--AGSPPRRGSPPLHAPPRRYPGPP-----------FDRSPERP----MNGYRSP 230
PRPAA G PP S P PP P PP +P RP R+P
Sbjct: 79 PRPAAPTGGPPPSPPSDPSPPPPPWPPPPPPLPPLFPPPP*STTAPPRP*SPSSGSSRTP 258
Query: 231 PPRLM--GRDGLRDYGPAAAALPPLRHEGRFP 260
P G G A++ PP R RFP
Sbjct: 259 TPASSSTGPPGPPSLTTRASSPPPRRA*PRFP 354
>TC228449 similar to GB|AAR99134.1|41058179|BT011476 RE11923p {Drosophila
melanogaster;} , partial (3%)
Length = 1112
Score = 43.5 bits (101), Expect = 2e-04
Identities = 60/200 (30%), Positives = 79/200 (39%), Gaps = 12/200 (6%)
Frame = +2
Query: 190 PAAAGSPPRRGSPP---LHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPA 246
P ++ SPP+ P L +P P PPF + + + S P +L R PA
Sbjct: 86 PPSSPSPPKP*PHPHINLVSPSGPTPNPPFSPTSKPSLAPRTSTPVKLQFPSRRRTRLPA 265
Query: 247 AAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDGF---SNE 303
AAA P R+ R P P L R D R + PPP + R R R +
Sbjct: 266 AAA--PSRN--RVPLPSLRPPR-----DQQRRPRRAQVPPPAEARARRRRRSVLHPQAPH 418
Query: 304 RKGFERRPLSPSAPLLPSLPPHRGGDRWSRDVRDRSRSPIRGGPPAKDYRRDPVMSRGGR 363
R+ RR L +P P RG D R R R+ + G P RR+P + RG
Sbjct: 419 RRAARRRRLE-----VPGGAPRRG------DRRRRGRARLLPGGP----RREPALLRGAL 553
Query: 364 DDR------RGGVGRDRIGG 377
+R R G G D G
Sbjct: 554 RERTADGPVRRGRGSDAAPG 613
>BG238374 similar to GP|11994340|db zinc finger protein-like; Ser/Thr protein
kinase-like protein {Arabidopsis thaliana}, partial
(62%)
Length = 470
Score = 43.5 bits (101), Expect = 2e-04
Identities = 28/63 (44%), Positives = 32/63 (50%), Gaps = 1/63 (1%)
Frame = +2
Query: 124 RPGGRPFG-RAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLNFARRDFC 182
R G +G R D G+G R G GL G + VRP GDW C A CG NFA R C
Sbjct: 137 RCGDSKYGDRVVDFGGFGGRGGSSFGLTGSD---VRP--GDWYCAAANCGAHNFASRSSC 301
Query: 183 NQC 185
+C
Sbjct: 302 FKC 310
Score = 33.5 bits (75), Expect = 0.16
Identities = 14/26 (53%), Positives = 16/26 (60%)
Frame = +2
Query: 160 REGDWMCPDALCGNLNFARRDFCNQC 185
R GDW C C +LNF RRD C +C
Sbjct: 71 RPGDWNCRS--CQHLNFQRRDSCQRC 142
>TC206638 similar to UP|Q9FYA8 (Q9FYA8) SC35-like splicing factor SCL30a, 30a
kD, partial (56%)
Length = 1050
Score = 42.0 bits (97), Expect = 5e-04
Identities = 34/113 (30%), Positives = 43/113 (37%), Gaps = 2/113 (1%)
Frame = +3
Query: 187 RPRPAAAGSPPRRG--SPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYG 244
R +P + RRG S +PPR P + RSP RSPPPR R RDY
Sbjct: 411 RKKPTEMRTRERRGRFSDRRRSPPRYSRSPRYSRSPRYS----RSPPPRHRSRSHSRDY- 575
Query: 245 PAAAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGR 297
P R R P R + + F R PP + +R R +
Sbjct: 576 ----YSPKRREYSRSVSPEGRRYSRERSYSQHNRERSFSRSPPYNGGSRSRSQ 722
Score = 28.1 bits (61), Expect = 6.8
Identities = 35/135 (25%), Positives = 49/135 (35%), Gaps = 8/135 (5%)
Frame = +3
Query: 51 RRDPPPVN----YSRSDRYSDDAGYRFRAGSSSPVHH---RRDADHRFPSDYNHFPRNRG 103
RR PP + YSRS RYS R R+ S S ++ RR+ + + R R
Sbjct: 468 RRSPPRYSRSPRYSRSPRYSRSPPPRHRSRSHSRDYYSPKRREYSRSVSPEGRRYSRERS 647
Query: 104 FG-GGRDFGRFRDPPHPYARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREG 162
+ R+ R PP+ G R ++ P G R L+ R R
Sbjct: 648 YSQHNRERSFSRSPPY-----NGGSRSRSQS---PAKGPGRSRSPSLNRDEREPARGRSP 803
Query: 163 DWMCPDALCGNLNFA 177
D C L +A
Sbjct: 804 SQ*AWDKTCPGLLYA 848
>TC226921 similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein precursor,
partial (5%)
Length = 641
Score = 42.0 bits (97), Expect = 5e-04
Identities = 43/153 (28%), Positives = 52/153 (33%), Gaps = 1/153 (0%)
Frame = +3
Query: 9 SPPSHHHHHQPLLSSLVVRPSLTENSPPGAAAHSNDYEPGELRRDPPPVNYSRSDRYSDD 68
SPPSHHHH Q L H PGE RR P R R
Sbjct: 120 SPPSHHHHRQTL-----------------RRRHHRLRRPGEARRPRP-----RQRR---- 221
Query: 69 AGYRFRAGSSSPVHHRRDADHRFPSDYNHFPRNRGFGGGRDFGRFRDPP-HPYARGRPGG 127
+R + + H R +H +H P R G PP H + RP
Sbjct: 222 --HRLQGPPQDHLRHLRAENHPLRR*RHHAP-------PRLLGNIHPPPRHRLSPRRPLP 374
Query: 128 RPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPR 160
R + R G G+HGR +P RPR
Sbjct: 375 RLLRESL------RRRGDPHGVHGRRHPRDRPR 455
Score = 30.4 bits (67), Expect = 1.4
Identities = 43/170 (25%), Positives = 55/170 (32%), Gaps = 11/170 (6%)
Frame = +2
Query: 186 KRPRPAAAGSP-PRRGSPPLHAPPRRYPGPP--FDRSPE--------RPMNGYRSPPPRL 234
+ P+ AA SP P PP PP P PP + SP + R PPP
Sbjct: 86 RSPQSAAPASPFPSLPPPPSPNPPPATPSPPPTWRSSPSSATATAAPSTRSATRPPPPPT 265
Query: 235 MGRDGLRDYGPAAAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRD 294
+ P AA P +H P + R PP W
Sbjct: 266 R*KSSTPTLTPPRAAAPSRKHPSSAAPPTVPTSSASTAP---------SRIPPATW---- 406
Query: 295 RGRDGFSNERKGFERRPLSPSAPLLPSLPPHRGGDRWSRDVRDRSRSPIR 344
R +S R P P+A S P R +SR R +R+ R
Sbjct: 407 --RSSWSTWTAAPSRPPSPPAARSRKSDSP-RWRVTYSRGSRTSTRATSR 547
>TC219587 similar to PIR|T49181|T49181 cyclophylin-like protein - Arabidopsis
thaliana {Arabidopsis thaliana;} , partial (19%)
Length = 1344
Score = 41.2 bits (95), Expect = 8e-04
Identities = 51/189 (26%), Positives = 64/189 (32%), Gaps = 4/189 (2%)
Frame = +1
Query: 190 PAAAGSPPR----RGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGP 245
P+ G P R RG +A RRY P +RSP G R+ R Y
Sbjct: 526 PSPNGMPKRIKKGRGFTERYAFARRYRTPSPERSPRTYRYGDRNIRRNFDRNTSYRSYSG 705
Query: 246 AAAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDGFSNERK 305
+ PP R F P R R Y R R P + +RDR R +
Sbjct: 706 RS---PPRR----FRSPPGGRNRPRYQSRRSRSRSNSRSPVRGHFSDRDRSRSPRRSPSP 864
Query: 306 GFERRPLSPSAPLLPSLPPHRGGDRWSRDVRDRSRSPIRGGPPAKDYRRDPVMSRGGRDD 365
R P+S R G R + DR RS + R P + R D
Sbjct: 865 EDRRPPISDQL-------KSRLGPRSDERLPDRGRSKSKSRSNGSSCSRSPDATPPKRYD 1023
Query: 366 RRGGVGRDR 374
+R V R R
Sbjct: 1024KRTSVSRSR 1050
>TC226883 similar to GB|AAO63346.1|28950845|BT005282 At1g67325 {Arabidopsis
thaliana;} , partial (40%)
Length = 784
Score = 40.4 bits (93), Expect = 0.001
Identities = 18/44 (40%), Positives = 25/44 (55%), Gaps = 2/44 (4%)
Frame = +3
Query: 160 REGDWMCPDALCGNLNFARRDFCN--QCKRPRPAAAGSPPRRGS 201
R+ DW CP CGN+NF+ R CN +C P+P + S + S
Sbjct: 189 RDNDWTCPK--CGNVNFSFRTVCNMRKCNTPKPGSQASKSDKNS 314
Score = 29.3 bits (64), Expect = 3.1
Identities = 15/46 (32%), Positives = 21/46 (45%), Gaps = 2/46 (4%)
Frame = +3
Query: 154 NPNVRPREGDWMCPDALCGNLNFARRDFCNQ--CKRPRPAAAGSPP 197
N + EG W C C N+N+ R CN+ C +PA + P
Sbjct: 309 NSKQKMPEGSWKCEK--CNNINYPFRTKCNRQNCGADKPAESEESP 440
>TC207021 similar to UP|Q940Z6 (Q940Z6) At1g12810/F13K23_4, partial (51%)
Length = 710
Score = 40.0 bits (92), Expect = 0.002
Identities = 20/54 (37%), Positives = 25/54 (46%), Gaps = 9/54 (16%)
Frame = +3
Query: 188 PRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPM---------NGYRSPPP 232
P+P +PP G PP PP Y G P + P+RP NG+ PPP
Sbjct: 141 PQPGYPSAPPHEGYPPPPPPPHGYGGYPPPQPPQRPPYDSYQGYFDNGHPPPPP 302
Score = 32.7 bits (73), Expect = 0.28
Identities = 29/90 (32%), Positives = 36/90 (39%), Gaps = 14/90 (15%)
Frame = +3
Query: 196 PPRRGSPPLHAPPRR-YPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPAAAALPPLR 254
PP GSP + PP+ YP P P GY PPP G YG PP R
Sbjct: 111 PPGYGSP--YPPPQPGYPSAP-------PHEGYPPPPPPPHG------YGGYPPPQPPQR 245
Query: 255 -----HEGRF--------PDPHLHRERMDY 271
++G F P PH H + +D+
Sbjct: 246 PPYDSYQGYFDNGHPPPPPPPHYHYQHVDH 335
>TC231484 similar to UP|Q6ZLD4 (Q6ZLD4) Cyclophylin-like protein, partial
(35%)
Length = 884
Score = 39.7 bits (91), Expect = 0.002
Identities = 52/194 (26%), Positives = 73/194 (36%), Gaps = 8/194 (4%)
Frame = +2
Query: 62 SDRYSDDAGYRFRAGSSSPVHHRRDADHRFPSDYNHFPRNRGFGGGRDFGRFRDPPHPYA 121
++RY+ YR + SP H R D +++ R + G RFR PP
Sbjct: 56 TERYAFARRYRTPSPERSPPHSYRYGDRNIRRNFDRNTSYRSYSGRSPPRRFRSPPG--G 229
Query: 122 RGRP---GGRPFGRAFDG-PGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNL--- 174
R RP R R+ G P G R R +P+ P + + D L L
Sbjct: 230 RNRPRYQSRRSRSRSISGSPVRGRFRERDRSRSPRRSPS--PEDRLPLISDRLKSRLGPR 403
Query: 175 -NFARRDFCNQCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPR 233
+ RD R + + GS R P PP+RY D+ P + RS
Sbjct: 404 SDEGLRDRGRSRSRSKSRSKGSSHSRS--PDATPPKRY-----DKRTSVPRS--RSRSSS 556
Query: 234 LMGRDGLRDYGPAA 247
G+ GL YG A+
Sbjct: 557 ASGQKGLVSYGDAS 598
Score = 38.9 bits (89), Expect = 0.004
Identities = 55/182 (30%), Positives = 66/182 (36%), Gaps = 6/182 (3%)
Frame = +2
Query: 199 RGSPPLHAPPRRYPGPPFDRSPER-PMNGYRSPPPRLMGR-DGLRDYGPAAAALPPLRHE 256
RG +A RRY P SPER P + YR + D Y + PP R
Sbjct: 47 RGFTERYAFARRYRTP----SPERSPPHSYRYGDRNIRRNFDRNTSYRSYSGRSPPRR-- 208
Query: 257 GRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDGFSNERKGFERRPLSPS- 315
F P R R Y R R P + RDR R RR SP
Sbjct: 209 --FRSPPGGRNRPRYQSRRSRSRSISGSPVRGRFRERDRSRS---------PRRSPSPED 355
Query: 316 -APLLPSLPPHRGGDRWSRDVRDRSRSPIRGGPPAK--DYRRDPVMSRGGRDDRRGGVGR 372
PL+ R G R +RDR RS R +K + R P + R D+R V R
Sbjct: 356 RLPLISDRLKSRLGPRSDEGLRDRGRSRSRSKSRSKGSSHSRSPDATPPKRYDKRTSVPR 535
Query: 373 DR 374
R
Sbjct: 536 SR 541
>AI442620
Length = 289
Score = 39.7 bits (91), Expect = 0.002
Identities = 16/27 (59%), Positives = 18/27 (66%)
Frame = +1
Query: 163 DWMCPDALCGNLNFARRDFCNQCKRPR 189
DWMC +CG +NFARR C QC PR
Sbjct: 166 DWMC--TICGYINFARRTSCYQCNEPR 240
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.143 0.479
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,419,292
Number of Sequences: 63676
Number of extensions: 544415
Number of successful extensions: 9889
Number of sequences better than 10.0: 653
Number of HSP's better than 10.0 without gapping: 6266
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8572
length of query: 379
length of database: 12,639,632
effective HSP length: 99
effective length of query: 280
effective length of database: 6,335,708
effective search space: 1773998240
effective search space used: 1773998240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC122722.5


