

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122722.4 + phase: 0
(307 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
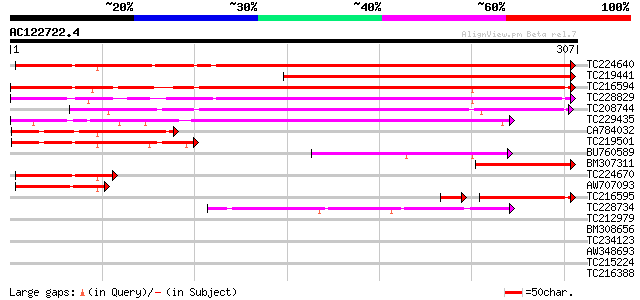
Score E
Sequences producing significant alignments: (bits) Value
TC224640 similar to UP|Q9XE35 (Q9XE35) ESTD24835(R2634) correspo... 339 8e-94
TC219441 weakly similar to UP|Q9LV42 (Q9LV42) Emb|CAB53482.1, pa... 234 3e-62
TC216594 similar to UP|Q9FMN3 (Q9FMN3) Emb|CAB53482.1, partial (... 221 4e-58
TC228829 similar to UP|Q9FMN3 (Q9FMN3) Emb|CAB53482.1, partial (... 220 7e-58
TC208744 similar to UP|Q9FMN3 (Q9FMN3) Emb|CAB53482.1, partial (... 195 2e-50
TC229435 weakly similar to UP|Q14883 (Q14883) Intestinal mucin (... 162 1e-40
CA784032 103 7e-23
TC219501 weakly similar to UP|Q9XE35 (Q9XE35) ESTD24835(R2634) c... 101 5e-22
BU760589 94 6e-20
BM307311 84 8e-17
TC224670 similar to UP|Q9XE35 (Q9XE35) ESTD24835(R2634) correspo... 71 7e-13
AW707093 47 1e-05
TC216595 40 3e-05
TC228734 weakly similar to UP|Q99738 (Q99738) Pinin, partial (4%) 45 5e-05
TC212979 35 0.001
BM308656 39 0.004
TC234123 37 0.009
AW348693 weakly similar to GP|19110824|gb ABC transporter ABCA.5... 36 0.025
TC215224 33 0.16
TC216388 similar to UP|Q8H1B5 (Q8H1B5) Hin1-like protein, partia... 33 0.21
>TC224640 similar to UP|Q9XE35 (Q9XE35) ESTD24835(R2634) corresponds to a
region of the predicted gene, partial (46%)
Length = 1605
Score = 339 bits (870), Expect = 8e-94
Identities = 179/309 (57%), Positives = 232/309 (74%), Gaps = 6/309 (1%)
Frame = +2
Query: 4 KTDSDVTSFDPSSP-RSQKQTPYYVQSPSRESSHDGDKSSTMQP-----SPMDSPSHQSY 57
K++SD+TS PSSP RS K+ YYVQSPSR+S HDGDKSS+MQ SPM+SPSH S+
Sbjct: 404 KSESDITSLAPSSPSRSPKRPVYYVQSPSRDS-HDGDKSSSMQATPISNSPMESPSHPSF 580
Query: 58 GHHSRASSSSRISGGYNSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGDFDGDGNGVSRR 117
G HSR SS+SR SG + SS GRK +RK NDKGW VI EE G Y +F G +RR
Sbjct: 581 GRHSRNSSASRFSGIFRSSS-GRKGSRKRNDKGWPECDVILEE-GSYHEFQD--KGFTRR 748
Query: 118 VQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKMLTV 177
Q +AV+ FV+VF++FC IIW AS YK +++VKSLTVHNFY GEGSD TGV TKMLTV
Sbjct: 749 FQALIAVLTFVVVFTVFCLIIWGASRPYKAEIAVKSLTVHNFYVGEGSDFTGVLTKMLTV 928
Query: 178 NCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQGSKV 237
N ++RM ++NPATFFGI+V S +NL++S++TVATGELKK++Q RKS R VSVN++G+KV
Sbjct: 929 NGTLRMSIYNPATFFGIHVHSTPINLVFSEITVATGELKKHYQPRKSHRIVSVNLEGTKV 1108
Query: 238 PLYGAGADVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAVDPHNNKYIK 297
PLYGAG+ + S ++ LTL FE++SRGNVVGKLV+T+H + ++C + ++ +K IK
Sbjct: 1109PLYGAGSTITVSQTGVEVPLTLNFEIRSRGNVVGKLVKTRHRKEITCPLVLNSSRSKPIK 1288
Query: 298 LKENECRYN 306
K+N C Y+
Sbjct: 1289FKKNSCTYD 1315
>TC219441 weakly similar to UP|Q9LV42 (Q9LV42) Emb|CAB53482.1, partial (15%)
Length = 890
Score = 234 bits (598), Expect = 3e-62
Identities = 115/158 (72%), Positives = 134/158 (84%)
Frame = +1
Query: 149 LSVKSLTVHNFYFGEGSDVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDL 208
L +S TVHNF FGEG D+TGVPTKMLTVNCS+RM VHNPATFFGI+VSSK+VNLMYS++
Sbjct: 199 LFFQSFTVHNFLFGEGLDLTGVPTKMLTVNCSVRMTVHNPATFFGIHVSSKAVNLMYSEM 378
Query: 209 TVATGELKKYHQQRKSRRTVSVNIQGSKVPLYGAGADVASSVDNGKISLTLVFEVKSRGN 268
TVATGELKK++ RKS RTVSVN+QGSKV LYGA A + VDNGKI +TLVFEV S GN
Sbjct: 379 TVATGELKKHYLSRKSTRTVSVNLQGSKVSLYGADASLTGLVDNGKIPMTLVFEVGSLGN 558
Query: 269 VVGKLVRTKHTQRVSCSIAVDPHNNKYIKLKENECRYN 306
+VG+LVR+KH +RVSCS+A+D HN + IKLKEN C YN
Sbjct: 559 IVGRLVRSKHRRRVSCSVAIDSHNIEPIKLKENACTYN 672
>TC216594 similar to UP|Q9FMN3 (Q9FMN3) Emb|CAB53482.1, partial (79%)
Length = 1430
Score = 221 bits (562), Expect = 4e-58
Identities = 136/313 (43%), Positives = 193/313 (61%), Gaps = 7/313 (2%)
Frame = +1
Query: 1 MHIKTDSDVTSFDPSSP-RSQKQTP-YYVQSPSRESSHDGDKSST--MQPSPMDSPSHQS 56
MH KTDS+VTS SSP RS + P YYVQSPSR+S HDG+K++T +P+ SPS
Sbjct: 115 MHAKTDSEVTSLAASSPTRSPPRRPLYYVQSPSRDS-HDGEKTATTSFHSTPVLSPSASP 291
Query: 57 YGHHSRASSSSRISGGYNSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGDFDGDGNGVSR 116
HSR SSS+R S +S L K W VIEEE G D NG+ R
Sbjct: 292 --PHSRHSSSTRFSKKDHSHSL----------KPWKQIDVIEEE--GLLQGDDRRNGLPR 429
Query: 117 RVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKMLT 176
R F VVGF+++FS F I+W AS KP++++KS+ + GSD TGV T M+T
Sbjct: 430 RCYFLAFVVGFLVLFSFFSLILWGASRPMKPKINIKSIKFDHVRVQAGSDATGVATDMIT 609
Query: 177 VNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQGSK 236
+N +++ N TFFG++V+S V L YSD+ +A G +KK++Q R+S+R +SV++ G+K
Sbjct: 610 LNSTLKFTYRNTGTFFGVHVTSTPVELSYSDIVIAAGNMKKFYQSRRSQRLLSVSVMGNK 789
Query: 237 VPLYGAGADVASS--VDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAVDPHN-N 293
+PLYG+GA ++S+ V + L L F ++SR V+GKLV+ K+ + + CSI +DP N
Sbjct: 790 IPLYGSGASLSSTTGVPTLPVPLNLNFVLRSRAYVLGKLVKPKYYKTIKCSITLDPKKLN 969
Query: 294 KYIKLKENECRYN 306
I LK++ C Y+
Sbjct: 970 AAISLKKS-CTYD 1005
>TC228829 similar to UP|Q9FMN3 (Q9FMN3) Emb|CAB53482.1, partial (75%)
Length = 1178
Score = 220 bits (560), Expect = 7e-58
Identities = 136/315 (43%), Positives = 187/315 (59%), Gaps = 9/315 (2%)
Frame = +2
Query: 1 MHIKTDSDVTSFDPSSP-RSQKQTPYYVQSPSRESSHDGDKS-----STMQPSPMDSPSH 54
MH KTDS+VTS D SS RS ++ YYVQSPS HDG+K+ ST SPM SP H
Sbjct: 23 MHAKTDSEVTSLDASSSTRSPRRAVYYVQSPS----HDGEKTTTSLHSTPVLSPMGSPPH 190
Query: 55 QSYGHHSRASSSSRISGGYNSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGDFDGDGNGV 114
SSSSR S + + N+K W G VIEEE + D + +
Sbjct: 191 SH-------SSSSRFSASRHRN--------NHNNKSWKGIDVIEEEGLLQSELDRQ-HSL 322
Query: 115 SRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKM 174
SRR F ++GF L+FSLF I+W AS KP++ +KS+ + GSD +GV T M
Sbjct: 323 SRRYYFLAFLLGFFLLFSLFSLILWCASRPMKPKILIKSIKFDHLRVQAGSDSSGVATDM 502
Query: 175 LTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQG 234
+T+N +++ N TFFG++V+S +L YSD+ +ATG LKK++Q RKS+R VSV + G
Sbjct: 503 ITMNSTVKFTYRNTGTFFGVHVTSTPFDLSYSDIVIATGNLKKFYQSRKSQRLVSVAVMG 682
Query: 235 SKVPLYGAGADVASS--VDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAVDPHN 292
+K+PLYG GA ++SS V + L L F ++SR V+G+LV+ K+ +RV CSI +DP
Sbjct: 683 NKIPLYGGGASLSSSTGVPTLPVPLNLTFVIRSRAYVLGRLVKPKYYKRVQCSINLDPKK 862
Query: 293 -NKYIKLKENECRYN 306
N I LK + C Y+
Sbjct: 863 INVPISLK-HSCTYD 904
>TC208744 similar to UP|Q9FMN3 (Q9FMN3) Emb|CAB53482.1, partial (38%)
Length = 1210
Score = 195 bits (495), Expect = 2e-50
Identities = 123/288 (42%), Positives = 173/288 (59%), Gaps = 15/288 (5%)
Frame = +1
Query: 33 ESSHDGDKSS-TMQPSPMDSP--------SHQSYGHHSRASSSSRISGGYNSSFLGRKVN 83
+SSHDG+K++ + SP+ SP S+ S GHHSR S+S+R SG SS G N
Sbjct: 4 DSSHDGEKTTNSFHSSPLQSPLGSPPHSHSNSSLGHHSRESASTRFSGSRKSSSSGN--N 177
Query: 84 RKVNDKGWIGS-KVIEEENGGYGDFDGDGNGVSRRVQFFVAVVGFVLVFSLFCFIIWAAS 142
RK + W IEEE G D + G R F V+GFVL+FS F I+W AS
Sbjct: 178 RKGPWRPWKDQFHAIEEE--GLIDAHDNARGFPRCCYFPAFVIGFVLLFSAFSLILWGAS 351
Query: 143 LHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVN 202
KP +S+KS+T F G+D++GV T ++++N S++M N ATFFG++V+S V+
Sbjct: 352 RPQKPAISLKSITFDQFGIQAGADMSGVATSLVSMNSSVKMTFRNTATFFGVHVTSTPVD 531
Query: 203 LMYSDLTVATGELKKYHQQRKSRRTVSVNIQGSKVPLYGAGADVASSVDNGK----ISLT 258
L Y LT+ATG + K++Q RKS+R+V V + GS +PLYG GA++ S NGK + LT
Sbjct: 532 LNYYQLTLATGTMPKFYQSRKSQRSVRVMVIGSHIPLYGGGANLNSV--NGKPVEPVPLT 705
Query: 259 LVFEVKSRGNVVGKLVRTKHTQRVSCSIAVDPHN-NKYIKLKENECRY 305
L V+SR V+GKLV+ K +++ CSI +DP K I L + +C Y
Sbjct: 706 LSVMVRSRAYVLGKLVKPKFYKKIECSIVMDPKKMGKAISLVK-KCTY 846
>TC229435 weakly similar to UP|Q14883 (Q14883) Intestinal mucin (Fragment),
partial (26%)
Length = 1075
Score = 162 bits (411), Expect = 1e-40
Identities = 109/294 (37%), Positives = 173/294 (58%), Gaps = 21/294 (7%)
Frame = +2
Query: 1 MHIKTDSDVTS--FDPSSP-RSQKQTP-YYVQSPSRESSHDGDKSSTMQPSPMDSPSHQS 56
MH K+DS+VTS + SSP RS + P YYVQSPS +HD +K S SPM SP H
Sbjct: 65 MHTKSDSEVTSNSMEQSSPSRSPPRRPLYYVQSPS---NHDVEKMS-YGSSPMGSPHHHF 232
Query: 57 YG------HHSRASSSSRISGG-------YNSSFLGRKVNRKVNDKGWIGSKVIEEENGG 103
+ HHSR SS+SR S ++SS +K++ N + +++
Sbjct: 233 HYYLSSPIHHSRESSTSRFSASLKNPRSNFSSSSSWKKLHPHPNPDAGL------DDDDD 394
Query: 104 YGDFDGDGNGVSRRVQ-FFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFG 162
D D D N R ++ +F + F+L F+LF I+W S YKP++ VKS+ N
Sbjct: 395 DDDGDDDLNHFPRNLRLYFCFFLLFLLFFTLFSLILWGTSKSYKPRIIVKSIVFENLNVQ 574
Query: 163 EGSDVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQR 222
G+D TGVPT ML++N ++R+L NPATFFG++V+S ++L Y L +A+G+++K++Q R
Sbjct: 575 SGNDGTGVPTDMLSLNSTVRILYRNPATFFGVHVTSTPLHLSYYQLAIASGQMQKFYQSR 754
Query: 223 KSRRTVSVNIQGSKVPLYGAGADVASSVDN-GKISLTLVFEVKS--RGNVVGKL 273
KS+R ++V + G ++PLYG + + ++ ++ ++L L ++ S +G +GK+
Sbjct: 755 KSQRKLAVVVLGHQIPLYGGVSVLGNTKEHLENVALPLKTDICSEIKGFHLGKI 916
>CA784032
Length = 423
Score = 103 bits (258), Expect = 7e-23
Identities = 63/97 (64%), Positives = 68/97 (69%), Gaps = 7/97 (7%)
Frame = +3
Query: 2 HIKTDSDVTSFDPSSPRSQKQTPYYVQSPSRESSHDGDKSSTMQP------SPMDSPSHQ 55
H KTDSDVTS D SS S K+ YYVQSPSR+S GDKSST SP+DSPSH
Sbjct: 126 HAKTDSDVTSMDTSS--SPKRAVYYVQSPSRDSH--GDKSSTATHATPACNSPVDSPSHH 293
Query: 56 SYGHHSRASSSSRISGG-YNSSFLGRKVNRKVNDKGW 91
SYGHHSRASSSSR+SGG YN + GRKV RK N GW
Sbjct: 294 SYGHHSRASSSSRVSGGSYNIASWGRKVTRK-NKLGW 401
>TC219501 weakly similar to UP|Q9XE35 (Q9XE35) ESTD24835(R2634) corresponds
to a region of the predicted gene, partial (16%)
Length = 544
Score = 101 bits (251), Expect = 5e-22
Identities = 68/116 (58%), Positives = 76/116 (64%), Gaps = 15/116 (12%)
Frame = +3
Query: 2 HIKTDSDVTSFDPSSPRSQKQTPYYVQSPSRESSHDGDKSSTMQP------SPMDSPSHQ 55
H KTDSDVTS D SS S K+ YYVQSPSR+S HDGDKSST SP+DSPSH
Sbjct: 207 HAKTDSDVTSMDTSS--SPKRAVYYVQSPSRDS-HDGDKSSTATHATPACNSPVDSPSHH 377
Query: 56 SYGHHSRASSSSRIS-GGYN--SSFLGRKVNRKVNDKGWIGS------KVIEEENG 102
SY HHSRASSSSR+S G YN +S+ GR N K K +GS KVI+EE G
Sbjct: 378 SYVHHSRASSSSRVSAGSYNNIASYWGR--NNKGTRKNKLGSWTHDQCKVIQEEEG 539
>BU760589
Length = 444
Score = 94.4 bits (233), Expect = 6e-20
Identities = 52/136 (38%), Positives = 78/136 (57%), Gaps = 27/136 (19%)
Frame = +1
Query: 164 GSDVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATG---------- 213
GSD +GV T M+T+N +++ N TFFG++V+S +L YSD+ +ATG
Sbjct: 37 GSDSSGVATDMITMNSTVKFTYRNTGTFFGVHVTSTPFDLSYSDIVIATGNVSIYTQIDH 216
Query: 214 ---------------ELKKYHQQRKSRRTVSVNIQGSKVPLYGAGADVASS--VDNGKIS 256
+LKK++Q RKS+R VSV + G+K+PLYG GA ++SS V +
Sbjct: 217 IINLII*FDLN*CTTQLKKFYQSRKSQRLVSVAVMGNKIPLYGGGASLSSSTGVPTLPVP 396
Query: 257 LTLVFEVKSRGNVVGK 272
L L F ++SR V+G+
Sbjct: 397 LNLTFVIRSRAYVLGR 444
>BM307311
Length = 371
Score = 84.0 bits (206), Expect = 8e-17
Identities = 38/54 (70%), Positives = 46/54 (84%)
Frame = +3
Query: 253 GKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAVDPHNNKYIKLKENECRYN 306
GKI +TLVF+V+SRGN+VGKLV +KH +RVSCS+A+D HN K IKLKEN C YN
Sbjct: 9 GKIPMTLVFDVRSRGNIVGKLVMSKHRRRVSCSVAIDSHNIKPIKLKENACTYN 170
>TC224670 similar to UP|Q9XE35 (Q9XE35) ESTD24835(R2634) corresponds to a
region of the predicted gene, partial (18%)
Length = 436
Score = 70.9 bits (172), Expect = 7e-13
Identities = 39/61 (63%), Positives = 47/61 (76%), Gaps = 6/61 (9%)
Frame = +2
Query: 4 KTDSDVTSFDPSSP-RSQKQTPYYVQSPSRESSHDGDKSSTMQP-----SPMDSPSHQSY 57
K++SD+TS PSSP RS K+ YYVQSPSR+S HDGDKSS+MQ SPM+SPSH S+
Sbjct: 257 KSESDITSLAPSSPSRSPKRPVYYVQSPSRDS-HDGDKSSSMQATPISNSPMESPSHPSF 433
Query: 58 G 58
G
Sbjct: 434 G 436
>AW707093
Length = 382
Score = 47.0 bits (110), Expect = 1e-05
Identities = 27/57 (47%), Positives = 38/57 (66%), Gaps = 6/57 (10%)
Frame = +2
Query: 4 KTDSDVTSFDPSSPRSQKQTP-YYVQSPSRESSHDGDKSSTMQP-----SPMDSPSH 54
K+ S++TS PSSP P YY+QSPS++ SH+ +KSS++Q SP+ SPSH
Sbjct: 215 KSKSNITSLAPSSPSKSPXHPIYYIQSPSKD-SHNRNKSSSIQATPISNSPIKSPSH 382
>TC216595
Length = 604
Score = 40.4 bits (93), Expect(2) = 3e-05
Identities = 22/53 (41%), Positives = 34/53 (63%), Gaps = 1/53 (1%)
Frame = +3
Query: 255 ISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAVDPHN-NKYIKLKENECRYN 306
+ L L F ++SR V+GKLV+ K+ + + CSI +DP N I LK++ C Y+
Sbjct: 81 VLLNLNFVLRSRAYVLGKLVKPKYYKTIQCSITLDPKKLNAAISLKKS-CTYD 236
Score = 24.6 bits (52), Expect(2) = 3e-05
Identities = 8/14 (57%), Positives = 13/14 (92%)
Frame = +1
Query: 234 GSKVPLYGAGADVA 247
G+K+PLYG+GA ++
Sbjct: 13 GNKIPLYGSGASLS 54
>TC228734 weakly similar to UP|Q99738 (Q99738) Pinin, partial (4%)
Length = 1156
Score = 44.7 bits (104), Expect = 5e-05
Identities = 43/172 (25%), Positives = 72/172 (41%), Gaps = 6/172 (3%)
Frame = +3
Query: 108 DGDGNGVSRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSD- 166
D + + RRV F A++GF+L+ L F+IW KP+ ++ TV+ F D
Sbjct: 237 DEERRQLPRRV--FAAILGFILLILLVIFLIWIILRPTKPRFILQDATVYAFNLSSTGDT 410
Query: 167 ---VTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMY--SDLTVATGELKKYHQQ 221
+T P LT+ + + NP G+Y + Y ++VAT L +Q
Sbjct: 411 PSPITPTP-NTLTLTMQVTLAAFNPNHRIGVYYTKLDAYAAYRGQQVSVAT-SLPATYQG 584
Query: 222 RKSRRTVSVNIQGSKVPLYGAGADVASSVDNGKISLTLVFEVKSRGNVVGKL 273
+ S + + VP+ + + K S ++ VK G V K+
Sbjct: 585 HRDTSVWSPYLYATAVPVSPFTLQI---LQQDKTSGGILVNVKVNGRVKWKV 731
>TC212979
Length = 811
Score = 35.0 bits (79), Expect(2) = 0.001
Identities = 20/44 (45%), Positives = 29/44 (65%)
Frame = +3
Query: 193 GIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQGSK 236
GI+V S VNL +SDL + LKK +Q R+S V VN++G++
Sbjct: 459 GIHVLSIPVNLDFSDLQLL--RLKKCYQPRRSHYIVPVNLEGNQ 584
Score = 24.6 bits (52), Expect(2) = 0.001
Identities = 10/12 (83%), Positives = 10/12 (83%)
Frame = +2
Query: 162 GEGSDVTGVPTK 173
GEGSD TGVP K
Sbjct: 380 GEGSDFTGVPRK 415
>BM308656
Length = 372
Score = 38.5 bits (88), Expect = 0.004
Identities = 23/57 (40%), Positives = 32/57 (55%), Gaps = 2/57 (3%)
Frame = -1
Query: 16 SPRSQKQTPYYVQSPSRESSHDGDK--SSTMQPSPMDSPSHQSYGHHSRASSSSRIS 70
SP+S+K T YY Q+PS SS + SS+ +P+P S +SY R S + R S
Sbjct: 282 SPKSRKHTAYYCQTPSLLSSREATSATSSSRKPAPPPRASPRSYSPPWRGSRTRRRS 112
>TC234123
Length = 753
Score = 37.4 bits (85), Expect = 0.009
Identities = 34/147 (23%), Positives = 65/147 (44%), Gaps = 1/147 (0%)
Frame = +3
Query: 128 VLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKMLTVNCSMRMLVHN 187
++ F + I++ + P + + +++ YF + G T L N S N
Sbjct: 189 LIFFGIATLILYLSMKPRNPTFDIPNASLNVVYFDSPQYLNGEFT--LLANFS------N 344
Query: 188 PATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQGSKVPL-YGAGADV 246
P G+ S ++ L YSD V++ +K + Q+ + R SVN+ S V L G +
Sbjct: 345 PNRRIGLRFESLNIELFYSDRLVSSQTIKPFTQRPRETRLQSVNLISSLVFLPQDVGVKL 524
Query: 247 ASSVDNGKISLTLVFEVKSRGNVVGKL 273
V+N +++ +GN+ G++
Sbjct: 525 QRQVEN------XXGQLQCKGNI*GEV 587
>AW348693 weakly similar to GP|19110824|gb ABC transporter ABCA.5
{Dictyostelium discoideum}, partial (1%)
Length = 699
Score = 35.8 bits (81), Expect = 0.025
Identities = 26/102 (25%), Positives = 46/102 (44%), Gaps = 2/102 (1%)
Frame = -2
Query: 186 HNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQGSKVPLY-GAGA 244
+NP IY + V++ Y D T+AT ++ + Q K+ + V + V LY
Sbjct: 548 YNPXXRVSIYYDTVEVSVRYEDQTLATNAVQPFFQSHKNVTRLHVGLTAQTVALYDSVPK 369
Query: 245 DVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKH-TQRVSCS 285
D+ +G I L + + R V + ++KH ++ CS
Sbjct: 368 DLRLERSSGDIELDVWMRARIRFKV--GVWKSKHRVLKIFCS 249
>TC215224
Length = 1219
Score = 33.1 bits (74), Expect = 0.16
Identities = 41/202 (20%), Positives = 83/202 (40%), Gaps = 7/202 (3%)
Frame = +2
Query: 113 GVSRRV--QFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGV 170
G R++ Q F + +++ L +IWA KP +++ +TV+ F
Sbjct: 134 GKKRKIFRQVFWCIEELLVLELLTILLIWAILRPTKPTFTLQDVTVYAF--------NAT 289
Query: 171 PTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMY--SDLTVATGELKKYHQQRKSRRTV 228
LT N + ++ NP G+Y + ++Y +T T + +Q
Sbjct: 290 IPNFLTSNFQVTLISRNPNDNIGVYYDRLEIYVIYRSQQITYRTA-IPPTYQGHNEINVW 466
Query: 229 SVNIQGSKVPLYGAG-ADVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCS-- 285
S + G+ +P+ ++ +G + +T+ + + R VG + ++ V C
Sbjct: 467 SPFVYGTNIPVAPFNFLRLSQDQSDGNVLVTIRADGRVRWK-VGAFISGRYHFYVRCPAF 643
Query: 286 IAVDPHNNKYIKLKENECRYNI 307
I+ P +N I + EN ++ I
Sbjct: 644 ISFGPRSNG-IVVGENAIKFQI 706
>TC216388 similar to UP|Q8H1B5 (Q8H1B5) Hin1-like protein, partial (43%)
Length = 1056
Score = 32.7 bits (73), Expect = 0.21
Identities = 36/172 (20%), Positives = 68/172 (38%), Gaps = 4/172 (2%)
Frame = +3
Query: 121 FVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKMLTVNCS 180
F ++ +++ + F+ W + V T+ F + P L + +
Sbjct: 213 FKLILTVIIIIGIAVFLFWLIVRPNVVKFHVTEATLTQFNY--------TPNNTLHYDLA 368
Query: 181 MRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQGSK-VPL 239
+ + V NP GIY M+ D + + ++Q KS ++ +G + VPL
Sbjct: 369 LNITVRNPNKRLGIYYDRIEARAMFHDARFDSQFPEPFYQGHKSTNVLNPVFKGQQLVPL 548
Query: 240 YGAGADVASSVDNGKISLTLVFEVKSRGNVVGKL--VRTKHTQ-RVSCSIAV 288
AD ++ + + +VK V KL +TK + +VSC + V
Sbjct: 549 ---NADQSAELKKENATGVYEIDVKMYLRVRFKLGVFKTKTLKPKVSCDLRV 695
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.132 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,115,019
Number of Sequences: 63676
Number of extensions: 212044
Number of successful extensions: 1796
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 1679
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1755
length of query: 307
length of database: 12,639,632
effective HSP length: 97
effective length of query: 210
effective length of database: 6,463,060
effective search space: 1357242600
effective search space used: 1357242600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC122722.4


