

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122169.1 + phase: 0 /pseudo
(459 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
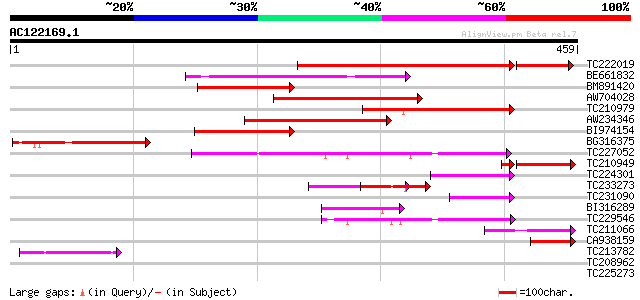
Score E
Sequences producing significant alignments: (bits) Value
TC222019 similar to UP|Q6K4N7 (Q6K4N7) Amino acid transporter-li... 261 1e-80
BE661832 135 3e-32
BM891420 similar to GP|20197120|gb| expressed protein {Arabidops... 115 4e-26
AW704028 114 7e-26
TC210979 109 3e-24
AW234346 99 3e-21
BI974154 99 4e-21
BG316375 86 3e-17
TC227052 weakly similar to GB|AAM19768.1|20453045|AY094388 AT3g5... 67 1e-11
TC210949 similar to UP|Q6K4N7 (Q6K4N7) Amino acid transporter-li... 59 5e-10
TC224301 weakly similar to UP|Q9MBG9 (Q9MBG9) Gb|AAC79623.2, par... 56 3e-08
TC233273 54 1e-07
TC231090 49 4e-06
BI316289 46 4e-05
TC229546 weakly similar to GB|AAM19768.1|20453045|AY094388 AT3g5... 43 3e-04
TC211066 42 6e-04
CA938159 similar to GP|20197120|gb| expressed protein {Arabidops... 42 7e-04
TC213782 similar to UP|Q6YU96 (Q6YU96) Amino acid transporter-li... 42 7e-04
TC208962 UP|Q9ARG2 (Q9ARG2) Amino acid transporter, complete 41 0.001
TC225273 similar to UP|Q6YKR7 (Q6YKR7) Cytochrome oxidase subuni... 36 0.031
>TC222019 similar to UP|Q6K4N7 (Q6K4N7) Amino acid transporter-like, partial
(84%)
Length = 979
Score = 261 bits (668), Expect(2) = 1e-80
Identities = 124/175 (70%), Positives = 142/175 (80%)
Frame = +1
Query: 234 VEFIIMEGDNLSGLFPGTSLHWGSLNLDGKHLFAILAALIILPTVWLKDLRFVSYLSAGG 293
+EFI +EGDNL+GLFPGTSL GS LD HLF ILAALII+PTVWLKDLR +S LSAGG
Sbjct: 19 LEFITLEGDNLTGLFPGTSLDLGSFRLDSVHLFGILAALIIIPTVWLKDLRIISILSAGG 198
Query: 294 VVGTALVGACVYAVGTRKDVGFHHTAPLVNWSGVPFAFGIYGFCFAGHSVFPNIYQSMAN 353
V T L+ CV+ VGT VGFHHT LVNWSG+P A GI+GFCFAGHSVFPNIYQSMA+
Sbjct: 199 VFATLLIVVCVFCVGTINGVGFHHTGQLVNWSGIPLAIGIHGFCFAGHSVFPNIYQSMAD 378
Query: 354 KKDFTKAMLICFVLPVFLYGSVGAAGFLMFGERTSSQITLDLPRDAFASKVSLWT 408
K+ FTKA++ICFVL + +YG V GFLMFG T SQITL++PRDAFASKV+LWT
Sbjct: 379 KRQFTKALIICFVLSITIYGGVAIMGFLMFGGETLSQITLNMPRDAFASKVALWT 543
Score = 57.0 bits (136), Expect(2) = 1e-80
Identities = 30/46 (65%), Positives = 34/46 (73%)
Frame = +2
Query: 411 HML***THWQEVWRNCCLIASHALIGASFFSEQHWLYLQFVLLF*F 456
+ML **THW+EV ++C I S LI SFF EQH LYL FVLLF*F
Sbjct: 566 NMLC**THWREVSKSCYQIESQVLIDVSFFLEQHLLYLPFVLLF*F 703
>BE661832
Length = 635
Score = 135 bits (341), Expect = 3e-32
Identities = 75/182 (41%), Positives = 101/182 (55%)
Frame = +1
Query: 143 TFTQTIFNGLNVLAGVGLLSAPDTVKQAGWASLLVIVVFAVVCFYTAELMRHCFQSREGI 202
+F T FNG+N + S P T+ GW SL ++ A FYT L++ I
Sbjct: 121 SFFHTCFNGINAV------SVPYTLASGGWLSLALLFAIAAATFYTGVLIKR*MDKDSNI 282
Query: 203 ISYPDIGEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGSLNLDG 262
+YPD+GE AFGK GR+ IS ++YTEL+ V F+I+EGDNLS LFP +H L + G
Sbjct: 283 RTYPDMGELAFGKTGRLIISGLIYTELFLVSVGFLILEGDNLSNLFPTVEIHTADLAIGG 462
Query: 263 KHLFAILAALIILPTVWLKDLRFVSYLSAGGVVGTALVGACVYAVGTRKDVGFHHTAPLV 322
K LF IL AL+ L +LR +SY+SA V +A++ + T VGFH L
Sbjct: 463 KKLFVILVALV------LDNLRILSYVSASRVFASAIIILSISWTATFDGVGFHQXGTLG 624
Query: 323 NW 324
NW
Sbjct: 625 NW 630
>BM891420 similar to GP|20197120|gb| expressed protein {Arabidopsis
thaliana}, partial (12%)
Length = 433
Score = 115 bits (288), Expect = 4e-26
Identities = 51/78 (65%), Positives = 67/78 (85%)
Frame = +2
Query: 153 NVLAGVGLLSAPDTVKQAGWASLLVIVVFAVVCFYTAELMRHCFQSREGIISYPDIGEAA 212
NV+AGVG+LS P T+K+AGW S++++V+FAV+C YTA LMR+CF+SREGI SYPDIGEAA
Sbjct: 176 NVMAGVGILSTPYTLKEAGWMSMVLMVLFAVICCYTATLMRYCFESREGITSYPDIGEAA 355
Query: 213 FGKYGRVFISIVLYTELY 230
FGKYGR+ +S+ L +Y
Sbjct: 356 FGKYGRIIVSVSLSIFIY 409
>AW704028
Length = 373
Score = 114 bits (286), Expect = 7e-26
Identities = 58/121 (47%), Positives = 75/121 (61%)
Frame = +3
Query: 214 GKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGSLNLDGKHLFAILAALI 273
GK GR+ IS +YTELY + F+I+EGDNL+ L P + + GK LF IL ALI
Sbjct: 6 GKIGRLIISGYMYTELYLVSIGFLILEGDNLNNLCPIEEVQIAGFVIGGKQLFVILVALI 185
Query: 274 ILPTVWLKDLRFVSYLSAGGVVGTALVGACVYAVGTRKDVGFHHTAPLVNWSGVPFAFGI 333
ILPTVWL +L +SY+SA GV +A++ + GT VGFH LVNW G+P A +
Sbjct: 186 ILPTVWLDNLSMLSYVSASGVFASAVIILSISWTGTFDGVGFHQKGTLVNWRGIPTAVSL 365
Query: 334 Y 334
Y
Sbjct: 366 Y 368
>TC210979
Length = 928
Score = 109 bits (272), Expect = 3e-24
Identities = 51/125 (40%), Positives = 78/125 (61%), Gaps = 2/125 (1%)
Frame = +2
Query: 286 VSYLSAGGVVGTALVGACVYAVGTRKDVGFHH--TAPLVNWSGVPFAFGIYGFCFAGHSV 343
+SY+SA GV+ T LV C++ VG + H T N + P A G+YG+C+AGH+V
Sbjct: 2 LSYISACGVIATILVVLCLFWVGLLDNADIHTQGTTKTFNLATFPVAIGLYGYCYAGHAV 181
Query: 344 FPNIYQSMANKKDFTKAMLICFVLPVFLYGSVGAAGFLMFGERTSSQITLDLPRDAFASK 403
F NIY +MAN+ F +L+CF + +Y +V G+ FG+ T SQ TL++P+ A+K
Sbjct: 182 FSNIYTAMANRNQFPGVLLVCFAICTSMYCAVAIMGYTAFGKATLSQYTLNMPQHLVATK 361
Query: 404 VSLWT 408
+++WT
Sbjct: 362 IAVWT 376
>AW234346
Length = 371
Score = 99.4 bits (246), Expect = 3e-21
Identities = 48/119 (40%), Positives = 74/119 (61%)
Frame = +2
Query: 191 LMRHCFQSREGIISYPDIGEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPG 250
L++ C I ++PDIG+ AFG GR+ +SI + +EL+ F+I+EGDNL+ L P
Sbjct: 11 LVKRCMDMDPDIKNFPDIGQRAFGDKGRIIVSIAMNSELFLVVTGFLILEGDNLNKLVPN 190
Query: 251 TSLHWGSLNLDGKHLFAILAALIILPTVWLKDLRFVSYLSAGGVVGTALVGACVYAVGT 309
L L + G +F ++AAL+ILPTV L+DL +SY+SA G + +++ +Y GT
Sbjct: 191 MQLELAGLTIGGTTIFTMIAALVILPTVLLEDLSMLSYVSASGALASSIFLLSIYWNGT 367
>BI974154
Length = 420
Score = 99.0 bits (245), Expect = 4e-21
Identities = 45/81 (55%), Positives = 57/81 (69%)
Frame = +2
Query: 150 NGLNVLAGVGLLSAPDTVKQAGWASLLVIVVFAVVCFYTAELMRHCFQSREGIISYPDIG 209
NG+NVL GVG+LS P K GW L ++V+FA++ FYT L+R C S + +YPDIG
Sbjct: 5 NGINVLCGVGILSTPYAAKVGGWLGLSILVIFAIISFYTGLLLRSCLDSEPELETYPDIG 184
Query: 210 EAAFGKYGRVFISIVLYTELY 230
+AAFG GR+ ISIVLY ELY
Sbjct: 185 QAAFGTTGRIAISIVLYVELY 247
>BG316375
Length = 427
Score = 85.9 bits (211), Expect = 3e-17
Identities = 49/117 (41%), Positives = 73/117 (61%), Gaps = 5/117 (4%)
Frame = +1
Query: 3 SNNSDKNKEKENDYFL--DANE---DETDIEAVNYDSDSSNNNDDDDEDDRIATHRPESF 57
+NNS+K KEK+ ++F D NE + D E +S+SS+++D+ ++ T R SF
Sbjct: 37 TNNSNK-KEKDLEFFFEEDINELGAQQMDAETAKLESESSSSDDEGND-----TTRRHSF 198
Query: 58 TSQQWPQSYNEALDPLTIAAAPNIGSVLRAPSVIYASFAAGRSSKSYLELQDGFLTG 114
TSQQWPQSY E D T+AA PN S+LR PS+IY+SF + + ++ + FL+G
Sbjct: 199 TSQQWPQSYKETTDSYTLAATPNFESILRVPSIIYSSFESRSKNNLDIDGKTPFLSG 369
Score = 35.8 bits (81), Expect = 0.041
Identities = 13/17 (76%), Positives = 15/17 (87%)
Frame = +1
Query: 326 GVPFAFGIYGFCFAGHS 342
G+P A GI+GFCFAGHS
Sbjct: 376 GIPLAIGIHGFCFAGHS 426
>TC227052 weakly similar to GB|AAM19768.1|20453045|AY094388
AT3g56200/F18O21_160 {Arabidopsis thaliana;} , partial
(68%)
Length = 1627
Score = 67.4 bits (163), Expect = 1e-11
Identities = 68/280 (24%), Positives = 119/280 (42%), Gaps = 21/280 (7%)
Frame = +3
Query: 148 IFNGLNVLAGVGLLSAPDTVKQAGWA-SLLVIVVFAVVCFYTAELMRHCFQSREGIISYP 206
+FN + G G++S P +K G + +I+V AV+ + + + S E +Y
Sbjct: 159 VFNVATSIVGAGIMSIPAIMKVLGVVPAFAMILVVAVLAELSVDFLMRFTHSGE-TTTYA 335
Query: 207 DIGEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLH------WGSLNL 260
+ AFG G + + + + ++I+ GD LSG G +H W ++
Sbjct: 336 GVMREAFGSGGALAAQVCVIITNVGGLILYLIIIGDVLSGKQNGGEVHLGILQQWFGIHW 515
Query: 261 DGKHLFAILAAL--IILPTVWLKDLRFVSYLSA-GGVVGTALVGACVYAVGTRKDVGFHH 317
FA+L L ++LP V K + + Y SA ++ A VG C T G
Sbjct: 516 WNSREFALLFTLVFVMLPLVLYKRVESLKYSSAVSTLLAVAFVGICCGLAITALVQGKTQ 695
Query: 318 TAPLVN-----------WSGVPFAFGIYGFCFAGHSVFPNIYQSMANKKDFTKAMLICFV 366
T L ++ VP + F F H I +A T A+ + +
Sbjct: 696 TPRLFPRLDYQTSFFDLFTAVPVVVTAFTFHFNVHP----IGFELAKASQMTTAVRLALL 863
Query: 367 LPVFLYGSVGAAGFLMFGERTSSQITLDLPRDAFASKVSL 406
L +Y ++G G+++FG+ T S I ++ ++A ++ SL
Sbjct: 864 LCAVIYLAIGLFGYMLFGDSTQSDILINFDQNAGSAVGSL 983
>TC210949 similar to UP|Q6K4N7 (Q6K4N7) Amino acid transporter-like, partial
(35%)
Length = 569
Score = 58.9 bits (141), Expect(2) = 5e-10
Identities = 32/48 (66%), Positives = 35/48 (72%)
Frame = +3
Query: 411 HML***THWQEVWRNCCLIASHALIGASFFSEQHWLYLQFVLLF*FHF 458
+ML **THWQ V R+CC I H IGAS F EQH L+LQFVL F*F F
Sbjct: 57 NMLC**THWQGV*RSCCQIEFHLHIGASCFLEQHLLHLQFVLPF*FPF 200
Score = 23.1 bits (48), Expect(2) = 5e-10
Identities = 9/10 (90%), Positives = 10/10 (100%)
Frame = +2
Query: 399 AFASKVSLWT 408
AFASKV+LWT
Sbjct: 5 AFASKVALWT 34
>TC224301 weakly similar to UP|Q9MBG9 (Q9MBG9) Gb|AAC79623.2, partial (29%)
Length = 667
Score = 56.2 bits (134), Expect = 3e-08
Identities = 25/68 (36%), Positives = 39/68 (56%)
Frame = +2
Query: 341 HSVFPNIYQSMANKKDFTKAMLICFVLPVFLYGSVGAAGFLMFGERTSSQITLDLPRDAF 400
H + Y SM +K F++ ICF + Y + G +LMFG+ SQ+TL+LP F
Sbjct: 2 HPIXXTXYNSMRDKSQFSRXXSICFSVCXLGYAAAGVLXYLMFGQEVESQVTLNLPTGKF 181
Query: 401 ASKVSLWT 408
+S V+++T
Sbjct: 182 SSHVAIFT 205
>TC233273
Length = 601
Score = 53.9 bits (128), Expect = 1e-07
Identities = 31/84 (36%), Positives = 46/84 (53%), Gaps = 2/84 (2%)
Frame = +1
Query: 243 NLSGLFPGTSLHWGSLNLDGKHLFAILAALIILPTVWLKDLRFVSYLSAGGVVGTALVGA 302
+LS LFP L+ G + L+ LFA++ L +LPTVWL+DL +SY+S G
Sbjct: 4 SLSSLFPSAHLNLGGIELNSHTLFAVITTLAVLPTVWLRDLSILSYIS----------GK 153
Query: 303 CVYAVGTRKDVGFHHTA--PLVNW 324
++ V R F+HT+ P + W
Sbjct: 154 SLFLVQIR----FYHTSWCPFL*W 213
Score = 49.7 bits (117), Expect = 3e-06
Identities = 23/56 (41%), Positives = 35/56 (62%)
Frame = +1
Query: 285 FVSYLSAGGVVGTALVGACVYAVGTRKDVGFHHTAPLVNWSGVPFAFGIYGFCFAG 340
F ++ AGGVV + LV C+ VG +DVGFH +N + +P G+YG+C++G
Sbjct: 436 FTMFVIAGGVVASILVVXCLLWVGI-EDVGFHSKGTTLNLATLPVDVGLYGYCYSG 600
>TC231090
Length = 559
Score = 49.3 bits (116), Expect = 4e-06
Identities = 20/52 (38%), Positives = 31/52 (59%)
Frame = +2
Query: 357 FTKAMLICFVLPVFLYGSVGAAGFLMFGERTSSQITLDLPRDAFASKVSLWT 408
F +L CF + LY G+ MFGE SQ TL++P++ A+K+++WT
Sbjct: 5 FPGVLLACFGICTLLYAGAAVLGYTMFGEAILSQFTLNMPKELVATKIAVWT 160
>BI316289
Length = 412
Score = 45.8 bits (107), Expect = 4e-05
Identities = 23/75 (30%), Positives = 42/75 (55%), Gaps = 8/75 (10%)
Frame = +3
Query: 253 LHWGSLNLDGKHLFAILAALIILPTVWLKDLRFVSYLSAGGVVGTALV--------GACV 304
L+ G + L+ + LFA++ L +LPTVWL+DL +SY+S + +V C+
Sbjct: 3 LNLGGIELNSRTLFAVITTLAVLPTVWLRDLSILSYISGKSLFLVQVVISYPLKLGPICL 182
Query: 305 YAVGTRKDVGFHHTA 319
+ G ++ F++T+
Sbjct: 183 WP*GHIAEIRFYYTS 227
>TC229546 weakly similar to GB|AAM19768.1|20453045|AY094388
AT3g56200/F18O21_160 {Arabidopsis thaliana;} , partial
(83%)
Length = 1336
Score = 42.7 bits (99), Expect = 3e-04
Identities = 45/171 (26%), Positives = 76/171 (44%), Gaps = 14/171 (8%)
Frame = +3
Query: 253 LHWGSLNLDGKHLFAILAAL--IILPTVWLKDLRFVSYLSA-GGVVGTALVGAC-VYAV- 307
+HW S FA+L L I+LP V + + + + SA ++ A V C V A+
Sbjct: 369 IHWWS-----SREFALLVVLFLILLPLVLYRRVESLKFSSAISTLLAVAFVTICTVLAIV 533
Query: 308 ----GTRKDVGF-----HHTAPLVNWSGVPFAFGIYGFCFAGHSVFPNIYQSMANKKDFT 358
G + HT+ ++ VP Y F F H I +A +
Sbjct: 534 AIVEGRTQSPRLIPCLDQHTSFFDLFTAVPVVVTAYTFHFNVHP----IGFELAKPSEMA 701
Query: 359 KAMLICFVLPVFLYGSVGAAGFLMFGERTSSQITLDLPRDAFASKVSLWTI 409
A+ I +L +Y S+G +G+L+FG+ T S I ++ ++A ++ SL +
Sbjct: 702 TAVRIALLLCCVIYFSIGLSGYLLFGDSTQSDILVNFDQNAGSALGSLLNV 854
>TC211066
Length = 805
Score = 42.0 bits (97), Expect = 6e-04
Identities = 24/74 (32%), Positives = 38/74 (50%)
Frame = +3
Query: 385 ERTSSQITLDLPRDAFASKVSLWTITHML***THWQEVWRNCCLIASHALIGASFFSEQH 444
E SQ TL++P++ A+ +++WT WQ VWR+ +LI SEQ
Sbjct: 42 EAILSQFTLNMPKELVATNIAVWTT---------WQ*VWRS*YRQTMPSLIYIPSSSEQG 194
Query: 445 WLYLQFVLLF*FHF 458
W +L ++L+ F F
Sbjct: 195 WFFLPYLLVSQFPF 236
>CA938159 similar to GP|20197120|gb| expressed protein {Arabidopsis
thaliana}, partial (9%)
Length = 422
Score = 41.6 bits (96), Expect = 7e-04
Identities = 22/37 (59%), Positives = 24/37 (64%)
Frame = +1
Query: 422 VWRNCCLIASHALIGASFFSEQHWLYLQFVLLF*FHF 458
V ++C I S LI SFF EQH LYL FVLLF F F
Sbjct: 1 VSKSCYQIESQVLIDVSFFLEQHLLYLPFVLLFLFLF 111
>TC213782 similar to UP|Q6YU96 (Q6YU96) Amino acid transporter-like protein,
partial (15%)
Length = 435
Score = 41.6 bits (96), Expect = 7e-04
Identities = 22/82 (26%), Positives = 43/82 (51%)
Frame = +1
Query: 9 NKEKENDYFLDANEDETDIEAVNYDSDSSNNNDDDDEDDRIATHRPESFTSQQWPQSYNE 68
N EN + ++++E++ + + +N N++D + + R S + WPQSY +
Sbjct: 175 NSVSENSFIIESDEEDEEKD-LNKGGVDGNDSDSSNYSNENPPQRKPSSYNISWPQSYRQ 351
Query: 69 ALDPLTIAAAPNIGSVLRAPSV 90
++D + +PNIG L PS+
Sbjct: 352 SIDLYSSVPSPNIG-YLGTPSL 414
>TC208962 UP|Q9ARG2 (Q9ARG2) Amino acid transporter, complete
Length = 1789
Score = 40.8 bits (94), Expect = 0.001
Identities = 56/283 (19%), Positives = 105/283 (36%), Gaps = 34/283 (12%)
Frame = +1
Query: 143 TFTQTIFNGLNVLAGVGLLSAPDTVKQAGW-ASLLVIVVFAVVCFYTAELMRHCFQSREG 201
TF + + + G G+LS V Q GW A +V+ +FAVV YT+ L+ C+++ +
Sbjct: 190 TFWMATAHIITAVIGSGVLSLAWAVAQLGWVAGPIVMFLFAVVNLYTSNLLTQCYRTGDS 369
Query: 202 I-----ISYPDIGEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWG 256
+ +Y + + G + Y L+ + + I ++ + H
Sbjct: 370 VSGHRNYTYMEAVNSILGGKKVKLCGLTQYINLFGVAIGYTIAASVSMMAIKRSNCYH-S 546
Query: 257 SLNLDGKHLFA------------ILAALIILPTVW----LKDLRFVSYLSAGGVVGTALV 300
S D H+ + I + + VW + + +Y S G +G A V
Sbjct: 547 SHGKDPCHMSSNGYMITFGIAEVIFSQIPDFDQVWWLSIVAAIMSFTYSSVGLSLGVAKV 726
Query: 301 -------GACV-YAVGTRKDVGFHHTAPLVNWSGVPFAFGIYGFCFAGHSVFPNIYQSM- 351
G+ + ++GT G + + W + A G F ++ + I ++
Sbjct: 727 AENKSFKGSLMGISIGTVTQAGTVTSTQKI-WRSLQ-ALGAMAFAYSFSIILIEIQDTIK 900
Query: 352 ---ANKKDFTKAMLICFVLPVFLYGSVGAAGFLMFGERTSSQI 391
A K KA + + Y G G+ FG+ +
Sbjct: 901 SPPAEHKTMRKATTLSIAVTTVFYLLCGCMGYAAFGDNAPGNL 1029
>TC225273 similar to UP|Q6YKR7 (Q6YKR7) Cytochrome oxidase subunit I
(Fragment), partial (10%)
Length = 1395
Score = 36.2 bits (82), Expect = 0.031
Identities = 16/54 (29%), Positives = 31/54 (56%), Gaps = 1/54 (1%)
Frame = +3
Query: 7 DKNKEKENDYFLDANEDETDIEAVNYDSDSSNNNDDDDEDDRIAT-HRPESFTS 59
D+N+ ++D + +ED +I A+ Y + +DDD+D+ +T H P + T+
Sbjct: 582 DENQRTDDDIESEMHEDTEEINALLYSESDGYSTEDDDDDEVTSTGHSPSTMTT 743
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.327 0.141 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,249,628
Number of Sequences: 63676
Number of extensions: 327727
Number of successful extensions: 3021
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 2477
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2747
length of query: 459
length of database: 12,639,632
effective HSP length: 101
effective length of query: 358
effective length of database: 6,208,356
effective search space: 2222591448
effective search space used: 2222591448
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122169.1


