

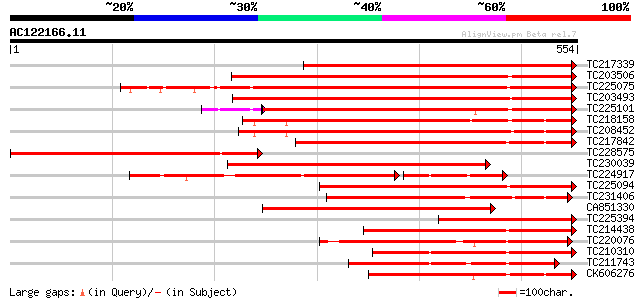
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122166.11 + phase: 0
(554 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase p... 482 e-136
TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (... 414 e-116
TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 413 e-116
TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like prote... 390 e-109
TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 337 2e-97
TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pe... 346 2e-95
TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (... 336 2e-92
TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 329 2e-90
TC228575 weakly similar to UP|O24298 (O24298) Pectinesterase pre... 324 6e-89
TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase i... 316 2e-86
TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4... 223 2e-84
TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 286 2e-77
TC231406 similar to UP|Q9FF78 (Q9FF78) Pectinesterase, partial (... 247 9e-66
CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.... 246 2e-65
TC225394 UP|SUSY_SOYBN (P13708) Sucrose synthase (Sucrose-UDP g... 245 4e-65
TC214438 similar to UP|Q708X4 (Q708X4) Pectin methylesterase (F... 236 2e-62
TC220076 homologue to UP|PME3_PHAVU (Q43111) Pectinesterase 3 pr... 236 2e-62
TC210310 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 234 8e-62
TC211743 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 233 2e-61
CK606276 222 3e-58
>TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precursor,
partial (48%)
Length = 923
Score = 482 bits (1240), Expect = e-136
Identities = 234/267 (87%), Positives = 246/267 (91%)
Frame = +1
Query: 288 KKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQA 347
+KTNVMLVGDG DAT+ITG+LNFIDGTTTFK+ATVAAVGDGFIAQDI FQNTAGPQKHQA
Sbjct: 1 EKTNVMLVGDGKDATVITGNLNFIDGTTTFKTATVAAVGDGFIAQDIWFQNTAGPQKHQA 180
Query: 348 VALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAA 407
VALRVGADQSVINRC+IDAFQDTLYAHSNRQFYRDS+ITGTVDFIFGNAAVVFQK L A
Sbjct: 181 VALRVGADQSVINRCRIDAFQDTLYAHSNRQFYRDSFITGTVDFIFGNAAVVFQKCDLVA 360
Query: 408 RKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTV 467
RKPM Q NMVTAQGREDPNQNT TSIQQC++ PSSDLKPV GSIKT+LGRPWKKYSRTV
Sbjct: 361 RKPMDKQNNMVTAQGREDPNQNTGTSIQQCNLTPSSDLKPVVGSIKTFLGRPWKKYSRTV 540
Query: 468 VLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAE 527
V+QS +D HIDP GWAEWDA SKDFLQTLYYGEYMN+G GAGT KRV WPGYHIIK AAE
Sbjct: 541 VMQSTLDSHIDPTGWAEWDAQSKDFLQTLYYGEYMNNGPGAGTSKRVNWPGYHIIKTAAE 720
Query: 528 ASKFTVTQLIQGNVWLKNTGVAFIEGL 554
ASKFTV QLIQGNVWLKNTGV FIEGL
Sbjct: 721 ASKFTVAQLIQGNVWLKNTGVNFIEGL 801
>TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (57%)
Length = 1320
Score = 414 bits (1065), Expect = e-116
Identities = 199/340 (58%), Positives = 250/340 (73%), Gaps = 2/340 (0%)
Frame = +2
Query: 217 GDFPSWVTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKK 276
G FP W+ +DRRLL + I+A++ V++ G+G KT+A+A+ AP++ R++IYV+
Sbjct: 5 GKFPGWLNKRDRRLLSLPLSAIQADITVSKTGNGTVKTIAEAIKKAPEHSTRRFIIYVRA 184
Query: 277 GTYKEN-IEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIR 335
G Y+EN +++G+KKTNVM +GDG T+ITG N IDG TTF SA+ AA G GFIA+DI
Sbjct: 185 GRYEENNLKVGRKKTNVMFIGDGKGKTVITGKKNVIDGMTTFHSASFAASGAGFIARDIT 364
Query: 336 FQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGN 395
F+N AGP KHQAVALRVGAD +V+ RC I +QD+ Y HSNRQF+R+ I GTVDFIFGN
Sbjct: 365 FENYAGPAKHQAVALRVGADHAVVYRCNIVGYQDSCYVHSNRQFFRECNIYGTVDFIFGN 544
Query: 396 AAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTY 455
AAVVFQK + ARKPMA QKN +TAQ R+DPNQNT S+ C ++P+ DL PV+GS TY
Sbjct: 545 AAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISLHNCRILPAPDLAPVKGSFPTY 724
Query: 456 LGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDF-LQTLYYGEYMNSGAGAGTGKRV 514
LGRPWK+YSRTV L S + HI P GW EW+ DF L TLYYGEYMN G GA G+RV
Sbjct: 725 LGRPWKQYSRTVYLVSYMGDHIHPRGWLEWNG---DFALNTLYYGEYMNYGPGAAVGQRV 895
Query: 515 TWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
WPGY +IK+ EA++FTV Q I G+ WL +TGVAF GL
Sbjct: 896 QWPGYRVIKSTMEANRFTVAQFISGSAWLPSTGVAFAAGL 1015
>TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (87%)
Length = 1744
Score = 413 bits (1062), Expect = e-116
Identities = 224/462 (48%), Positives = 296/462 (63%), Gaps = 16/462 (3%)
Frame = +1
Query: 109 IKRRVNSP----REEIALNDCEELMDLSMDRVWDSVLTLTKN---------NIDSQHDAH 155
I R SP R A+ DC +L+DLS D V L+ ++N N+ S D
Sbjct: 202 ILSRFGSPLANFRLSTAIADCLDLLDLSSD-VLSWALSASQNPKGKHNSTGNLSS--DLR 372
Query: 156 TWLSSVLTNHATCLDGLEGSSRVV---MESDLHDLISRARSSLAVLVSVLPPKANDGFID 212
TWLS+ L + TC++G EG++ +V + + + ++S LA VLP A D F
Sbjct: 373 TWLSAALAHPETCMEGFEGTNSIVKGLVSAGIGQVVSLVEQLLA---QVLP--AQDQFDA 537
Query: 213 EKLNGDFPSWVTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVI 272
G FPSW+ K+R+LL++ + +V VA DGSG + + AV +APD R+VI
Sbjct: 538 ASSKGQFPSWIKPKERKLLQAIA--VTPDVTVALDGSGNYAKIMDAVLAAPDYSMKRFVI 711
Query: 273 YVKKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQ 332
VKKG Y EN+EI KKK N+M++G GMDAT+I+G+ + +DG TTF+SAT A G GFIA+
Sbjct: 712 LVKKGVYVENVEIKKKKWNIMILGQGMDATVISGNRSVVDGWTTFRSATFAVSGRGFIAR 891
Query: 333 DIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFI 392
DI FQNTAGP+KHQAVALR +D SV RC I +QD+LY H+ RQF+RD I+GTVD+I
Sbjct: 892 DISFQNTAGPEKHQAVALRSDSDLSVFFRCGIFGYQDSLYTHTMRQFFRDCTISGTVDYI 1071
Query: 393 FGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSI 452
FG+A VFQ L +K + NQKN +TA GR+DPN+ T S Q C++ SDL P G+
Sbjct: 1072FGDATAVFQNCFLRVKKGLPNQKNTITAHGRKDPNEPTGFSFQFCNITADSDLIPSVGTA 1251
Query: 453 KTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGK 512
+TYLGRPWK YSRTV +QS + I GW EW+ L TLYY EYMN+GAGAG
Sbjct: 1252QTYLGRPWKSYSRTVFMQSYMSEVIGAEGWLEWN--GNFALDTLYYAEYMNTGAGAGVAN 1425
Query: 513 RVTWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
RV WPGYH + ++++AS FTV+Q I+GN+WL +TGV F GL
Sbjct: 1426RVKWPGYHALNDSSQASNFTVSQFIEGNLWLPSTGVTFTAGL 1551
>TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like protein
(At3g49220), partial (59%)
Length = 1298
Score = 390 bits (1002), Expect = e-109
Identities = 186/338 (55%), Positives = 242/338 (71%), Gaps = 1/338 (0%)
Frame = +1
Query: 218 DFPSWVTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKG 277
+FP+W+ +DRRLL + I+A++VV++DG+G KT+A+A+ P+ R +IY++ G
Sbjct: 136 NFPTWLNGRDRRLLSLPLSQIQADIVVSKDGNGTVKTIAEAIKKVPEYSSRRIIIYIRAG 315
Query: 278 TYKE-NIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRF 336
Y+E N+++G+KKTNVM +GDG T+ITG N+ TTF +A+ AA G GFIA+D+ F
Sbjct: 316 RYEEDNLKLGRKKTNVMFIGDGKGKTVITGGRNYYQNLTTFHTASFAASGSGFIAKDMTF 495
Query: 337 QNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNA 396
+N AGP +HQAVALRVGAD +V+ RC I +QDT+Y HSNRQFYR+ I GTVDFIFGNA
Sbjct: 496 ENYAGPGRHQAVALRVGADHAVVYRCNIIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNA 675
Query: 397 AVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYL 456
AVVFQ L ARKPMA QKN +TAQ R+DPNQNT SI C ++ + DL+ +GS TYL
Sbjct: 676 AVVFQNCTLWARKPMAQQKNTITAQNRKDPNQNTGISIHNCRIMATPDLEASKGSYPTYL 855
Query: 457 GRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTW 516
GRPWK Y+RTV + S + H+ P GW EW+ +S L T YYGEYMN G G+ G+RV W
Sbjct: 856 GRPWKLYARTVFMLSYIGDHVHPRGWLEWNTSS-FALDTCYYGEYMNYGPGSALGQRVNW 1032
Query: 517 PGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
GY I + EAS+FTV Q I G+ WL +TGVAFI GL
Sbjct: 1033AGYRAINSTVEASRFTVGQFISGSSWLPSTGVAFIAGL 1146
>TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (64%)
Length = 1177
Score = 337 bits (863), Expect(2) = 2e-97
Identities = 169/311 (54%), Positives = 218/311 (69%), Gaps = 4/311 (1%)
Frame = +3
Query: 248 GSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMDATIITGS 307
G + V AV +AP+ RYVI++K+G Y EN+EI KKK N+M+VGDGMDATII+G+
Sbjct: 207 GRENYTKVMDAVLAAPNYSMQRYVIHIKRGVYYENVEIKKKKWNLMMVGDGMDATIISGN 386
Query: 308 LNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAF 367
+FIDG TTF+SAT A G GFIA+DI FQNTAGP+KHQAVALR +D SV RC I +
Sbjct: 387 RSFIDGWTTFRSATFAVSGRGFIARDITFQNTAGPEKHQAVALRSDSDLSVFFRCGIFGY 566
Query: 368 QDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPN 427
QD+LY H+ RQFYR+ I+GTVDFIFG+A +FQ ++A+K + NQKN +TA GR++P+
Sbjct: 567 QDSLYTHTMRQFYRECKISGTVDFIFGDATAIFQNCHISAKKGLPNQKNTITAHGRKNPD 746
Query: 428 QNTATSIQQCDVIPSSDLKPVQGSIK---TYLGRPWKKYSRTVVLQSVVDGHIDPAGWAE 484
+ T SIQ C++ DL S TYLGRPWK YSRT+ +QS + + P GW E
Sbjct: 747 EPTGFSIQFCNISADYDLVNSVNSCNSTHTYLGRPWKPYSRTIFMQSYISDVLRPEGWLE 926
Query: 485 WDAASKDF-LQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGNVWL 543
W+ DF L TLYY EYMN G GAG RV W GYH++ ++++AS FTV+Q I+GN+ L
Sbjct: 927 WNG---DFALDTLYYAEYMNYGPGAGVANRVKWQGYHVMNDSSQASNFTVSQFIEGNLCL 1097
Query: 544 KNTGVAFIEGL 554
+TGV F GL
Sbjct: 1098PSTGVTFTAGL 1130
Score = 38.1 bits (87), Expect(2) = 2e-97
Identities = 22/64 (34%), Positives = 37/64 (57%)
Frame = +2
Query: 188 ISRARSSLAVLVSVLPPKANDGFIDEKLNGDFPSWVTSKDRRLLESSVGDIKANVVVAQD 247
I + S L L++ + P +D F G +PSWV + +R+LL+++V + + VV D
Sbjct: 35 IGQVMSLLQQLLTQVKP-VSDHFSFSSPQGQYPSWVKTGERKLLQANV--VSFDAVVTAD 205
Query: 248 GSGK 251
G+GK
Sbjct: 206 GTGK 217
>TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pectin
methylesterase-like protein, partial (59%)
Length = 1291
Score = 346 bits (887), Expect = 2e-95
Identities = 178/334 (53%), Positives = 228/334 (67%), Gaps = 7/334 (2%)
Frame = +3
Query: 228 RRLLESSVGD---IKANVVVAQDGSGKFKTVAQAVASAPDNGKTR---YVIYVKKGTYKE 281
R+LL++ VGD ++ V V+QDGSG F T+ A+A+AP+ + ++IYV G Y+E
Sbjct: 102 RKLLQAKVGDEVVVRDIVTVSQDGSGNFTTINDAIAAAPNKSVSTDGYFLIYVTAGVYEE 281
Query: 282 NIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAG 341
N+ I KKKT +M+VGDG++ TIITG+ + +DG TTF SAT+A VG GF+ ++ +NTAG
Sbjct: 282 NVSIDKKKTYLMMVGDGINKTIITGNRSVVDGWTTFSSATLAVVGQGFVGVNMTIRNTAG 461
Query: 342 PQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQ 401
KHQAVALR GAD S C + +QDTLY HS RQFY + I GTVDFIFGNA VVFQ
Sbjct: 462 AVKHQAVALRSGADLSTFYSCSFEGYQDTLYVHSLRQFYSECDIYGTVDFIFGNAKVVFQ 641
Query: 402 KSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWK 461
K+ R PM+ Q N +TAQGR DPNQ+T SI C + + DL G + TYLGRPWK
Sbjct: 642 NCKMYPRLPMSGQFNAITAQGRTDPNQDTGISIHNCTIRAADDLAASNG-VATYLGRPWK 818
Query: 462 KYSRTVVLQSVVDGHIDPAGWAEWDAASKDF-LQTLYYGEYMNSGAGAGTGKRVTWPGYH 520
+YSRTV +Q+V+D I GW EWD DF L TLYY EY NSG G+GT RVTWPGYH
Sbjct: 819 EYSRTVYMQTVMDSVIHAKGWREWDG---DFALSTLYYAEYSNSGPGSGTDNRVTWPGYH 989
Query: 521 IIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
+I NA +A+ FTV+ + G+ WL TGV++ L
Sbjct: 990 VI-NATDAANFTVSNFLLGDDWLPQTGVSYTNNL 1088
>TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (56%)
Length = 1273
Score = 336 bits (861), Expect = 2e-92
Identities = 173/337 (51%), Positives = 225/337 (66%), Gaps = 6/337 (1%)
Frame = +1
Query: 224 TSKDRRLLESSVGD---IKANVVVAQDGSGKFKTVAQAVASAPDNGKTR---YVIYVKKG 277
TS+ R+L+ S + V+V+ G + ++ A+A+AP+N K +++YV++G
Sbjct: 229 TSRTERILKESGSQGILLYDFVIVSHYGIDNYTSIGDAIAAAPNNTKPEDGYFLVYVREG 408
Query: 278 TYKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQ 337
Y+E + I K+K N++LVGDG++ TIITG+ + IDG TTF S+T A G+ FIA D+ F+
Sbjct: 409 LYEEYVVIPKEKKNILLVGDGINKTIITGNHSVIDGWTTFNSSTFAVSGERFIAVDVTFR 588
Query: 338 NTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAA 397
NTAGP+KHQAVA+R AD S RC + +QDTLY HS RQFYR+ I GTVDFIFGNAA
Sbjct: 589 NTAGPEKHQAVAVRNNADLSTFYRCSFEGYQDTLYVHSLRQFYRECEIYGTVDFIFGNAA 768
Query: 398 VVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLG 457
VVFQ K+ ARKP+ NQKN VTAQGR DPNQNT SIQ C + + DL S ++LG
Sbjct: 769 VVFQGCKIYARKPLPNQKNAVTAQGRTDPNQNTGISIQNCSIDAAPDLVADLNSTMSFLG 948
Query: 458 RPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWP 517
RPWK YSRTV LQS + I PAGW EW+ L TL+YGE+ N G G+ T RVTWP
Sbjct: 949 RPWKVYSRTVYLQSYIGNVIQPAGWLEWNGTVG--LDTLFYGEFNNYGPGSNTSNRVTWP 1122
Query: 518 GYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
GY ++ NA +A FTV GN WL +T + + EGL
Sbjct: 1123GYSLL-NATQAWNFTVLNFTLGNTWLPDTDIPYTEGL 1230
>TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (48%)
Length = 1225
Score = 329 bits (844), Expect = 2e-90
Identities = 163/275 (59%), Positives = 199/275 (72%)
Frame = +3
Query: 280 KENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNT 339
KENI+I + N+M+VGDGM ATI+TG+ N IDG+TTF+SAT A GDGFIA+DI F+NT
Sbjct: 6 KENIDIKRTVKNLMIVGDGMGATIVTGNHNAIDGSTTFRSATFAVDGDGFIARDITFENT 185
Query: 340 AGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVV 399
AGPQKHQAVALR GAD SV RC +QDTLY ++NRQFYRD I GTVDFIFG+A V
Sbjct: 186 AGPQKHQAVALRSGADHSVFYRCSFRGYQDTLYVYANRQFYRDCDIYGTVDFIFGDAVAV 365
Query: 400 FQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRP 459
Q + RKPM+NQ+N VTAQGR DPN+NT I C + + DLK VQGS +T+LGRP
Sbjct: 366 LQNCNIYVRKPMSNQQNTVTAQGRTDPNENTGIIIHNCRITAAGDLKAVQGSFRTFLGRP 545
Query: 460 WKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGY 519
W+KYSRTVV++S +DG I PAGW W + L TLYY E+ N+GAGA TG RV W G+
Sbjct: 546 WQKYSRTVVMKSALDGLISPAGWFPW--SGNFALSTLYYAEHANTGAGASTGGRVDWAGF 719
Query: 520 HIIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
+I ++ EA KFTV + G W+ +GV F EGL
Sbjct: 720 RVI-SSTEAVKFTVGNFLAGGSWIPGSGVPFDEGL 821
>TC228575 weakly similar to UP|O24298 (O24298) Pectinesterase precursor ,
partial (44%)
Length = 755
Score = 324 bits (831), Expect = 6e-89
Identities = 166/247 (67%), Positives = 202/247 (81%)
Frame = +2
Query: 1 MAIQQSLLDKPRKSIPKTFWLVLSLVAIISSSALIISHLNKPISFFNLSSAPNLCEHALD 60
MAIQQSLLD+PRKS+ KT L+ S+ A++ SSA + S+L K SFFN SS +LC+HALD
Sbjct: 17 MAIQQSLLDRPRKSVSKTICLIFSIAAVMISSAFVGSYLIKSTSFFNQSSPQHLCDHALD 196
Query: 61 TKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRVNSPREEI 120
+CLTHVSEVVQG L+ TKDHK + L S L K T+HI++ M+TA+ IK R+NSP+EE
Sbjct: 197 RATCLTHVSEVVQGPILTPTKDHKFNLLQSFLMKYTSHIKRVMNTASSIKLRINSPKEEE 376
Query: 121 ALNDCEELMDLSMDRVWDSVLTLTKNNIDSQHDAHTWLSSVLTNHATCLDGLEGSSRVVM 180
AL+DC ELMDLS+ RV DS++TLTK I+SQ DAHTWLSSVLTNHATCLDGLEGS+R M
Sbjct: 377 ALHDCVELMDLSISRVRDSMVTLTKQTIESQQDAHTWLSSVLTNHATCLDGLEGSARAFM 556
Query: 181 ESDLHDLISRARSSLAVLVSVLPPKANDGFIDEKLNGDFPSWVTSKDRRLLESSVGDIKA 240
+ +L DLISRAR+SLA+ V+VLPPK + IDE L+GDFPSWV+SKDRRLLES+VGDIKA
Sbjct: 557 KDELEDLISRARTSLAMFVAVLPPKV-EQIIDEPLSGDFPSWVSSKDRRLLESTVGDIKA 733
Query: 241 NVVVAQD 247
NVVVA+D
Sbjct: 734 NVVVAKD 754
>TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (70%)
Length = 913
Score = 316 bits (810), Expect = 2e-86
Identities = 151/256 (58%), Positives = 193/256 (74%)
Frame = +1
Query: 214 KLNGDFPSWVTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIY 273
K+ FP+W+++KDR+LL+++V + N++VA+DG+G F T+A+AVA AP++ TR+VI+
Sbjct: 145 KIKDGFPTWLSTKDRKLLQAAVNETNFNLLVAKDGTGNFTTIAEAVAVAPNSSATRFVIH 324
Query: 274 VKKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQD 333
+K G Y EN+E+ +KKTN+M VGDG+ T++ S N +DG TTF+SATVA VGDGFIA+
Sbjct: 325 IKAGAYFENVEVIRKKTNLMFVGDGIGKTVVKASRNVVDGWTTFQSATVAVVGDGFIAKG 504
Query: 334 IRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIF 393
I F+N+AGP KHQAVALR G+D S +C A+QDTLY HS RQFYRD + GTVDFIF
Sbjct: 505 ITFENSAGPSKHQAVALRSGSDFSAFYKCSFVAYQDTLYVHSLRQFYRDCDVYGTVDFIF 684
Query: 394 GNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIK 453
GNAA V Q L ARKP NQ+N+ TAQGREDPNQNT SI C V ++DL PV+ K
Sbjct: 685 GNAATVLQNCNLYARKPNENQRNLFTAQGREDPNQNTGISILNCKVAAAADLIPVKSQFK 864
Query: 454 TYLGRPWKKYSRTVVL 469
YLGRPWKKYSRTV L
Sbjct: 865 NYLGRPWKKYSRTVYL 912
>TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4L23.27
{Arabidopsis thaliana;} , partial (69%)
Length = 1483
Score = 223 bits (568), Expect(2) = 2e-84
Identities = 131/269 (48%), Positives = 164/269 (60%), Gaps = 5/269 (1%)
Frame = +2
Query: 118 EEIALNDCEELMDLSMDRVWDSVLTLTKNNIDSQHDAHTWLSSVLTNHATCLDG-----L 172
E+ A DC +L + ++ R+ ++ TK N Q D TWLS+ LTN TC +G +
Sbjct: 389 EKAAWADCLQLYEYTIQRLNKTINPNTKCN---QTDTQTWLSTALTNLETCKNGFYELGV 559
Query: 173 EGSSRVVMESDLHDLISRARSSLAVLVSVLPPKANDGFIDEKLNGDFPSWVTSKDRRLLE 232
+M +++ L+S S PP +GF P+WV DR+LL+
Sbjct: 560 PDYVLPLMSNNVTKLLSNTLSLNKGPYQYKPPSYKEGF---------PTWVKPGDRKLLQ 712
Query: 233 SSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNV 292
SS ANVVVA+DGSGK+ TV AV +AP + RYVIYVK G Y E +E+ K N+
Sbjct: 713 SSSVASNANVVVAKDGSGKYTTVKAAVDAAPKSSSGRYVIYVKSGVYNEQVEV--KGNNI 886
Query: 293 MLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRV 352
MLVGDG+ TIITGS + GTTTF+SATVAAVGDGFIAQDI F+NTAG HQAVA R
Sbjct: 887 MLVGDGIGKTIITGSKSVGGGTTTFRSATVAAVGDGFIAQDITFRNTAGAANHQAVAFRS 1066
Query: 353 GADQSVINRCKIDAFQDTLYAHSNRQFYR 381
G+D SV RC + FQDTLY HS RQFYR
Sbjct: 1067GSDLSVFYRCSFEGFQDTLYVHSERQFYR 1153
Score = 108 bits (270), Expect(2) = 2e-84
Identities = 56/102 (54%), Positives = 67/102 (64%)
Frame = +3
Query: 385 ITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSD 444
I GTVDFIFGNAA V Q + AR P + VTAQGR DPNQNT I V +S
Sbjct: 1164 IYGTVDFIFGNAAAVLQNCNIYARTP-PQRTITVTAQGRTDPNQNTGIIIHNSKVTGASG 1340
Query: 445 LKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWD 486
P S+K+YLGRPW+KYSRTV +++ +D I+PAGW EWD
Sbjct: 1341 FNP--SSVKSYLGRPWQKYSRTVFMKTYLDSLINPAGWMEWD 1460
>TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (47%)
Length = 919
Score = 286 bits (732), Expect = 2e-77
Identities = 141/252 (55%), Positives = 176/252 (68%)
Frame = +3
Query: 303 IITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRC 362
II+G+ + +DG TTF+SAT A G GFIA+DI FQNTAGP+KHQAVALR D SV RC
Sbjct: 3 IISGNRSVVDGWTTFRSATFAVSGRGFIARDISFQNTAGPEKHQAVALRSDTDLSVFFRC 182
Query: 363 KIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQG 422
I +QD+LY H+ RQF+R+ ITGTVD+IFG+A VFQ L +K + NQKN +TA G
Sbjct: 183 GIFGYQDSLYTHTMRQFFRECTITGTVDYIFGDATAVFQNCFLRVKKGLPNQKNTITAHG 362
Query: 423 REDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGW 482
R+DPN+ T S Q C++ SDL P S ++YLGRPWK YSRTV +QS + I GW
Sbjct: 363 RKDPNEPTGFSFQFCNITADSDLVPWVSSTQSYLGRPWKSYSRTVFMQSYMSEVIRGEGW 542
Query: 483 AEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGNVW 542
EW+ L+TLYYGEYMN+GAGAG RV WPGYH ++ +AS FTV Q I+GN+W
Sbjct: 543 LEWN--GNFALETLYYGEYMNTGAGAGLANRVKWPGYHPFNDSNQASNFTVAQFIEGNLW 716
Query: 543 LKNTGVAFIEGL 554
L +TGV + GL
Sbjct: 717 LPSTGVTYTAGL 752
>TC231406 similar to UP|Q9FF78 (Q9FF78) Pectinesterase, partial (35%)
Length = 851
Score = 247 bits (631), Expect = 9e-66
Identities = 127/241 (52%), Positives = 154/241 (63%)
Frame = +2
Query: 310 FIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQD 369
F+DGT TF +AT A G FIA+D+ F+NTAGPQK QAVAL ADQ+V RC+IDAFQD
Sbjct: 2 FVDGTPTFSTATFAVFGRNFIARDMGFRNTAGPQKQQAVALMTSADQAVYYRCQIDAFQD 181
Query: 370 TLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQN 429
+LYAHSNRQFYR+ I GTVDFIFGN+AVV Q + R PM Q+N +TAQG+ DPN N
Sbjct: 182 SLYAHSNRQFYRECNIYGTVDFIFGNSAVVLQNCNIMPRVPMQGQQNPITAQGKTDPNMN 361
Query: 430 TATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAAS 489
T SIQ C++ P DL S+KTYLGRPWK YS V + S + I P GW W S
Sbjct: 362 TGISIQNCNITPFGDL----SSVKTYLGRPWKNYSTPVFM*SPMGIFIHPNGWLPWVGNS 529
Query: 490 KDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVA 549
T++Y E+ N G GA T RV W G +I +AS FTV + G W+ +G
Sbjct: 530 AP--DTIFYAEFQNVGPGASTKNRVNWKGLRVI-TRKQASMFTVKAFLSGERWITASGAP 700
Query: 550 F 550
F
Sbjct: 701 F 703
>CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.150 -
Arabidopsis thaliana, partial (37%)
Length = 700
Score = 246 bits (627), Expect = 2e-65
Identities = 129/228 (56%), Positives = 158/228 (68%), Gaps = 1/228 (0%)
Frame = +2
Query: 248 GSGKFKTVAQAV-ASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMDATIITG 306
GSG FKTV A+ A+A KTR+VI+VKKG Y+ENIE+ N+MLVGDG+ TIIT
Sbjct: 14 GSGNFKTVQDALNAAAKRKEKTRFVIHVKKGVYRENIEVALHNDNIMLVGDGLRNTIITS 193
Query: 307 SLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDA 366
+ + DG TT+ SAT G FIA+DI FQN+AG K QAVALR +D SV RC I
Sbjct: 194 ARSVQDGYTTYSSATAGIDGLHFIARDITFQNSAGVHKGQAVALRSASDLSVFYRCGIMG 373
Query: 367 FQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDP 426
+QDTL AH+ RQFYR YI GTVDFIFGNAAVVFQ + AR+P+ Q NM+TAQGR DP
Sbjct: 374 YQDTLMAHAQRQFYRQCYIYGTVDFIFGNAAVVFQNCYIFARRPLEGQANMITAQGRGDP 553
Query: 427 NQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVD 474
QNT SI + + DLKPV T+LGRPW++YSR VV+++ +D
Sbjct: 554 FQNTGISIHNSQIRAAPDLKPVVDKYNTFLGRPWQQYSRVVVMKTFMD 697
>TC225394 UP|SUSY_SOYBN (P13708) Sucrose synthase (Sucrose-UDP
glucosyltransferase) (Nodulin-100) , complete
Length = 3328
Score = 245 bits (625), Expect = 4e-65
Identities = 115/135 (85%), Positives = 120/135 (88%)
Frame = -2
Query: 420 AQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDP 479
AQGREDPNQNT TSIQQC++ PSSDLKPV GSIKT GRPWKKYSRTVV+QS +D HIDP
Sbjct: 3327 AQGREDPNQNTGTSIQQCNLTPSSDLKPVVGSIKTXXGRPWKKYSRTVVMQSTLDSHIDP 3148
Query: 480 AGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQG 539
GWAEWDA SKDFLQTLYYGEYMN+G GAGT KRV WPGYHIIK AAEASKFTV QLIQG
Sbjct: 3147 TGWAEWDAQSKDFLQTLYYGEYMNNGPGAGTSKRVNWPGYHIIKTAAEASKFTVAQLIQG 2968
Query: 540 NVWLKNTGVAFIEGL 554
NVWLKNTGV FIEGL
Sbjct: 2967 NVWLKNTGVNFIEGL 2923
>TC214438 similar to UP|Q708X4 (Q708X4) Pectin methylesterase (Fragment) ,
partial (49%)
Length = 1026
Score = 236 bits (603), Expect = 2e-62
Identities = 120/209 (57%), Positives = 143/209 (68%)
Frame = +3
Query: 346 QAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKL 405
QAVALRVG D S C I AFQDTLY H+NRQF+ I GTVDFIFGN+AVVFQ +
Sbjct: 6 QAVALRVGGDLSAFFNCDILAFQDTLYVHNNRQFFEKCLIAGTVDFIFGNSAVVFQDCDI 185
Query: 406 AARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSR 465
AR P + QKNMVTAQGR DPNQNT IQ+C + ++DL+ V+ + KTYLGRPWK+YSR
Sbjct: 186 HARLPSSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATNDLESVKKNFKTYLGRPWKEYSR 365
Query: 466 TVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNA 525
TV++QS + IDP GW EW + L TL Y EY N+G GAGT RVTW GY +I +A
Sbjct: 366 TVIMQSSISDVIDPIGWHEW--SGNFALSTLVYREYQNTGPGAGTSNRVTWKGYKVITDA 539
Query: 526 AEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
AEA +T I G+ WL +TG F GL
Sbjct: 540 AEARDYTPGSFIGGSSWLGSTGFPFSLGL 626
>TC220076 homologue to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precursor
(Pectin methylesterase 3) (PE 3) , partial (39%)
Length = 808
Score = 236 bits (602), Expect = 2e-62
Identities = 123/250 (49%), Positives = 154/250 (61%), Gaps = 2/250 (0%)
Frame = +2
Query: 303 IITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRC 362
++TGS+ F A G GFIA+DI F N AG KHQAVA R G+D+SV RC
Sbjct: 50 VLTGSMLFF----------AAVKGKGFIAKDIGFVNNAGASKHQAVAFRSGSDRSVFFRC 199
Query: 363 KIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQG 422
+ FQDTLYAHSNRQFYRD ITGT+DFIFGNAA VFQ K+ R+P+ NQ N +TAQG
Sbjct: 200 SFNGFQDTLYAHSNRQFYRDCDITGTIDFIFGNAAAVFQNCKIMPRQPLPNQFNTITAQG 379
Query: 423 REDPNQNTATSIQQCDVIPSSDLKPVQGSI--KTYLGRPWKKYSRTVVLQSVVDGHIDPA 480
++D NQNT IQ+ S P++ ++ TYLGRPWK +S TV++QS + + P
Sbjct: 380 KKDRNQNTGIIIQK------SKFTPLENNLTAPTYLGRPWKDFSTTVIMQSDIGSFLKPV 541
Query: 481 GWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGN 540
GW W + + + T++Y EY N+G GA +RV W GY EA KFTV IQG
Sbjct: 542 GWMSW-VPNVEPVSTIFYAEYQNTGPGADVSQRVKWAGYKPTLTDGEAGKFTVQSFIQGP 718
Query: 541 VWLKNTGVAF 550
WL N V F
Sbjct: 719 EWLPNAAVQF 748
>TC210310 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (38%)
Length = 678
Score = 234 bits (597), Expect = 8e-62
Identities = 114/200 (57%), Positives = 140/200 (70%)
Frame = +1
Query: 355 DQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQ 414
D SV +C + +QDTLY HS RQFYR+ I GTVDFIFGNAAVV Q + AR P N+
Sbjct: 1 DLSVFYKCSFEGYQDTLYVHSERQFYRECNIYGTVDFIFGNAAVVLQNCNIFARNP-PNK 177
Query: 415 KNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVD 474
N +TAQGR DPNQNT SI V +SDL+PVQ S++TYLGRPWK+YSRTV +++ +D
Sbjct: 178 VNTITAQGRTDPNQNTGISIHNSRVTAASDLRPVQNSVRTYLGRPWKQYSRTVFMKTYLD 357
Query: 475 GHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVT 534
G I+PAGW EW + L TLYYGEYMN+G G+ T +RV W GY +I +A+EASKF+V
Sbjct: 358 GLINPAGWMEW--SGNFALDTLYYGEYMNTGPGSSTARRVKWSGYRVITSASEASKFSVA 531
Query: 535 QLIQGNVWLKNTGVAFIEGL 554
I GN WL +T V F L
Sbjct: 532 NFIAGNAWLPSTKVPFTPSL 591
>TC211743 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (39%)
Length = 601
Score = 233 bits (594), Expect = 2e-61
Identities = 118/206 (57%), Positives = 144/206 (69%)
Frame = +2
Query: 332 QDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDF 391
QDI F+NTAG HQAVALR G+D SV RC + +QDTLY +S+RQFYR+ I GTVDF
Sbjct: 2 QDITFRNTAGATNHQAVALRSGSDLSVFYRCSFEGYQDTLYVYSDRQFYRECDIYGTVDF 181
Query: 392 IFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGS 451
IFGNAAVVFQ + AR P N+ N +TAQGR DPNQNT SI V +SDL
Sbjct: 182 IFGNAAVVFQNCNIYARNP-PNKVNTITAQGRTDPNQNTGISIHNSKVTAASDLM----G 346
Query: 452 IKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTG 511
++TYLGRPW++YSRTV +++ +D I+P GW EW + L TLYYGEYMN+G G+ T
Sbjct: 347 VRTYLGRPWQQYSRTVFMKTYLDSLINPEGWLEW--SGNFALSTLYYGEYMNTGPGSSTA 520
Query: 512 KRVTWPGYHIIKNAAEASKFTVTQLI 537
RV W GYH+I +A+EASKFTV I
Sbjct: 521 NRVNWLGYHVITSASEASKFTVGNFI 598
>CK606276
Length = 751
Score = 222 bits (566), Expect = 3e-58
Identities = 116/208 (55%), Positives = 134/208 (63%), Gaps = 4/208 (1%)
Frame = +1
Query: 351 RVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKP 410
R GAD S C + +QDTLY HS RQFY+ I GTVDFIFGNAA + Q + R P
Sbjct: 1 RNGADMSTFYNCSFEGYQDTLYVHSLRQFYKSCDIYGTVDFIFGNAAALLQDCNMYPRLP 180
Query: 411 MANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGS---IKTYLGRPWKKYSRTV 467
M NQ N +TAQGR DPNQNT SIQ C +I +SDL + IKTYLGRPWK+YSRTV
Sbjct: 181 MQNQFNAITAQGRTDPNQNTGISIQNCCIIAASDLGDATNNYNGIKTYLGRPWKEYSRTV 360
Query: 468 VLQSVVDGHIDPAGWAEWDAASKDF-LQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAA 526
+QS +DG IDP GW EW S DF L TLYY E+ N G G+ T RV W GYH+I +
Sbjct: 361 YMQSFIDGLIDPKGWNEW---SGDFALSTLYYAEFANWGPGSNTSNRVAWEGYHLI-DEK 528
Query: 527 EASKFTVTQLIQGNVWLKNTGVAFIEGL 554
+A FTV + IQG WL TGV F GL
Sbjct: 529 DADDFTVHKFIQGEKWLPQTGVPFXAGL 612
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.131 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,749,260
Number of Sequences: 63676
Number of extensions: 277710
Number of successful extensions: 1500
Number of sequences better than 10.0: 121
Number of HSP's better than 10.0 without gapping: 1408
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1413
length of query: 554
length of database: 12,639,632
effective HSP length: 102
effective length of query: 452
effective length of database: 6,144,680
effective search space: 2777395360
effective search space used: 2777395360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122166.11


