

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122162.14 - phase: 0
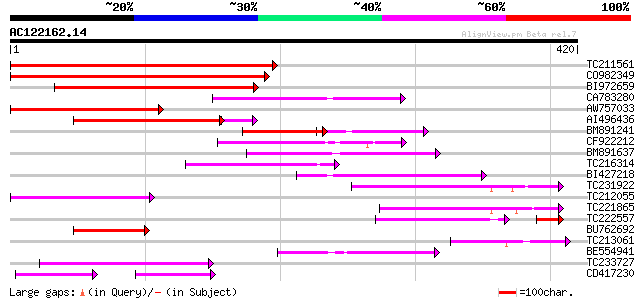
(420 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 186 2e-47
CO982349 174 7e-44
BI972659 142 2e-34
CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polym... 116 2e-26
AW757033 115 4e-26
AI496436 110 7e-26
BM891241 76 5e-22
CF922212 101 7e-22
BM891637 97 1e-20
TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {A... 97 2e-20
BI427218 90 2e-18
TC231922 72 6e-13
TC212055 71 8e-13
TC221865 65 7e-11
TC222557 55 1e-10
BU762692 60 2e-09
TC213061 56 3e-08
BE554941 55 6e-08
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 53 2e-07
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 37 6e-07
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 186 bits (471), Expect = 2e-47
Identities = 84/198 (42%), Positives = 128/198 (64%)
Frame = +1
Query: 1 MRNAVSRNLFKGFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKV 60
MR AV NL+K + + +G IS LQYADDT+ GEA+ +N+ +K +LR FE+ S L++
Sbjct: 316 MRRAVEENLYKPYLVGANGVPISILQYADDTIFFGEAAKENVEAIKVILRSFELVSNLRI 495
Query: 61 NFYKSCLMGINVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRK 120
NF KSC V ++ A ++L+CS PF YLG+P+GANS + W+ ++ + RK
Sbjct: 496 NFAKSCFGVFGVTDQWKQEAANYLHCSLLAFPFIYLGIPIGANSRRCHMWDSIIRKCERK 675
Query: 121 LHSWGNKYVSLGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKK 180
L W +++S GGR+ L+ +V+ SIPI++ SF ++P V K+VKI FLWGG KK
Sbjct: 676 LSKWK*RHISFGGRVTLIKSVLTSIPIYFFSFFRVPQSVVDKLVKI*RTFLWGGGPDQKK 855
Query: 181 LCWVKWRVVCQPRSKGGL 198
+ W++W +C P+ +GG+
Sbjct: 856 ISWIRWEKLCLPKERGGI 909
>CO982349
Length = 795
Score = 174 bits (441), Expect = 7e-44
Identities = 82/192 (42%), Positives = 122/192 (62%)
Frame = +3
Query: 1 MRNAVSRNLFKGFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKV 60
MR AV R F F + + +S LQYADDT+ EA+++N+ +K +LR FE+ASGLK+
Sbjct: 219 MREAVDRKRFNSFLVGKNKEPVSILQYADDTIFFEEATMENMKVIKTILRCFELASGLKI 398
Query: 61 NFYKSCLMGINVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRK 120
NF +S I P + A +FLNCS +PF YLG+P+ AN +P++ + K
Sbjct: 399 NFAESRFGAIWKPDHWCKEAAEFLNCSMLSMPFSYLGIPIEANPRCREI*DPIIRKCETK 578
Query: 121 LHSWGNKYVSLGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKK 180
L W +++SLGGR+ L+NA++ ++PI++ SF + P+KV +K+V IQ FLWGG ++
Sbjct: 579 LARWKQRHISLGGRVTLINAILTALPIYFFSFFRAPSKVVQKLVNIQRKFLWGGGSXQRR 758
Query: 181 LCWVKWRVVCQP 192
+ WVKW VC P
Sbjct: 759 IAWVKWETVCLP 794
>BI972659
Length = 453
Score = 142 bits (359), Expect = 2e-34
Identities = 62/151 (41%), Positives = 97/151 (64%)
Frame = +1
Query: 34 IGEASVDNLWTLKALLRGFEMASGLKVNFYKSCLMGINVPSEFMTMACDFLNCSEGVVPF 93
+GEAS DN+ LK++LRGFEM SGL++N+ KS + + A LNC + +PF
Sbjct: 1 VGEASWDNIIVLKSMLRGFEMVSGLRINYAKSQFGIVGFQPNWAHDAAQLLNCRQLDIPF 180
Query: 94 KYLGLPVGANSFKLVTWEPLLEQLSRKLHSWGNKYVSLGGRIVLLNAVINSIPIFYLSFL 153
YLG+P+ + + WEPL+ + KL W KY+S+GGR+ L+ +V+N++PI+ LSF
Sbjct: 181 HYLGMPIAVKASSRMVWEPLINKFKAKLSKWNQKYLSMGGRVTLIKSVLNALPIYLLSFF 360
Query: 154 KMPNKVWRKIVKIQCDFLWGGARGGKKLCWV 184
K+P ++ K+V +Q F+WGG + ++ WV
Sbjct: 361 KIPQRIVDKLVSLQRTFMWGGNQHHNRISWV 453
>CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polymerase
homolog T24M8.8 - Arabidopsis thaliana, partial (4%)
Length = 456
Score = 116 bits (290), Expect = 2e-26
Identities = 55/143 (38%), Positives = 79/143 (54%)
Frame = +2
Query: 151 SFLKMPNKVWRKIVKIQCDFLWGGARGGKKLCWVKWRVVCQPRSKGGLGVRDIRVVNLSL 210
SF +P K+ K+ +Q FLWGG +K+ WV W+ VC P++KGGLG++D+R N +L
Sbjct: 5 SFFSLP*KIIAKLESLQRRFLWGGEADSRKIAWVNWKTVCLPKAKGGLGIKDLRTFNTTL 184
Query: 211 LAKWRWRVLQGEEGLWKEVLIEKYGTRVCNLLVEDDGSWPSYISRWWKDVAFLEDEGGER 270
L KWRW + ++ W +VL KYG ++GS S S WWKD+ +
Sbjct: 185 LGKWRWDLFYIQQEPWAKVLQSKYG----GWRALEEGSSGSKDSAWWKDLIKTQQLQRNI 352
Query: 271 WFNAEVVRKVGCGNSTSFWKDPW 293
E + KVG G+ FW+D W
Sbjct: 353 PLKRETIWKVGGGDRIKFWEDLW 421
>AW757033
Length = 441
Score = 115 bits (288), Expect = 4e-26
Identities = 55/114 (48%), Positives = 79/114 (69%)
Frame = -2
Query: 1 MRNAVSRNLFKGFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKV 60
MR AV+RN +K F + +S LQYADDT+ GEA+++N+ +K +LRGFE+ASGLK+
Sbjct: 347 MREAVARNHYKSFAVGKYKKPVSILQYADDTIFFGEATMENVRVIKTILRGFELASGLKI 168
Query: 61 NFYKSCLMGINVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLL 114
NF KS I+VP + A +F+NCS +PF YLG+P+GAN + TW+P++
Sbjct: 167 NFAKSRFGAISVPD*WRKEAAEFMNCSLLSLPFSYLGIPIGANPRRRETWDPII 6
>AI496436
Length = 414
Score = 110 bits (276), Expect(2) = 7e-26
Identities = 54/112 (48%), Positives = 76/112 (67%)
Frame = +1
Query: 48 LLRGFEMASGLKVNFYKSCLMGINVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKL 107
+LR FE++SGLK+NF KS I + A D+LN S +PF YLG+P+GANS
Sbjct: 4 ILRCFELSSGLKINFAKSQFGTICKSDIWRKEAADYLNSSVLSMPFVYLGIPIGANSRHS 183
Query: 108 VTWEPLLEQLSRKLHSWGNKYVSLGGRIVLLNAVINSIPIFYLSFLKMPNKV 159
WEP++ + RKL SW KYVS GGR+ L+N+V+ ++PI+ LSF ++P+KV
Sbjct: 184 DVWEPVVRKCERKLASWKQKYVSFGGRVTLINSVLTALPIYLLSFFRIPDKV 339
Score = 24.6 bits (52), Expect(2) = 7e-26
Identities = 11/26 (42%), Positives = 15/26 (57%)
Frame = +2
Query: 158 KVWRKIVKIQCDFLWGGARGGKKLCW 183
K+ K+ IQ FLWGG +K+ W
Sbjct: 335 KL*TKLTIIQRRFLWGGDLE*RKIAW 412
>BM891241
Length = 407
Score = 75.9 bits (185), Expect(2) = 5e-22
Identities = 30/63 (47%), Positives = 42/63 (66%)
Frame = -2
Query: 173 GGARGGKKLCWVKWRVVCQPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIE 232
GG KK+ WVKW VVC P++KGGLG++DI N++LL KW W + ++ LW ++
Sbjct: 406 GGDHDHKKIPWVKWEVVCLPKTKGGLGIKDISKFNIALLGKWIWALASDQQQLWARIINS 227
Query: 233 KYG 235
KYG
Sbjct: 226 KYG 218
Score = 46.6 bits (109), Expect(2) = 5e-22
Identities = 25/84 (29%), Positives = 41/84 (48%), Gaps = 1/84 (1%)
Frame = -3
Query: 228 EVLIEKYGTRVCNLLVEDDGSWPSYISRWWKDVAFLEDEGGERWFNAEVVRKVGCGNSTS 287
E+LI+ + ++ G + S WW+D+ + F+ + KVGCG+ +
Sbjct: 240 ELLIQSMEAGLIFSIIVSKGGY----SYWWRDLRKFYHQSDHSIFHQYMSWKVGCGDKIN 73
Query: 288 FWKDPWRG-DIPFSRKYPRLFAIS 310
FW D W G D +KY +LF I+
Sbjct: 72 FWTDKWLGEDYTLEQKYNQLFLIN 1
>CF922212
Length = 445
Score = 101 bits (251), Expect = 7e-22
Identities = 58/143 (40%), Positives = 77/143 (53%), Gaps = 3/143 (2%)
Frame = -2
Query: 155 MPNKVWRKIVKIQCDFLWGGARGGKKLCWVKWRVVCQPRSKGGLGVRDIRVVNLSLLAKW 214
+PNKV K+V++Q FLWGG G KK+ WV W VC P+ K GLG RD+R N +LL K
Sbjct: 441 VPNKVLDKLVRLQQWFLWGGGLGQKKIAWVNWDSVCLPKEKEGLGNRDLRKFNYALLGKR 262
Query: 215 RWRVLQGEEGLWKEVLIEKYGTRVCNLLVEDDGSWPSYISRWWKDVAFL--EDEGGERWF 272
RW + + L VL KY R NL D+ S WW++++F+ E G WF
Sbjct: 261 RWNLFHHQGELGARVLDSKY-KRWRNL---DEERRVKSESFWWQEISFITHSTEDGS-WF 97
Query: 273 NAEV-VRKVGCGNSTSFWKDPWR 294
++GC FW+D WR
Sbjct: 96 EKRTKTGRLGCRAKVRFWEDGWR 28
>BM891637
Length = 427
Score = 97.4 bits (241), Expect = 1e-20
Identities = 48/146 (32%), Positives = 74/146 (49%), Gaps = 2/146 (1%)
Frame = -1
Query: 176 RGGKKLCWVKWRVVCQPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKY- 234
R GKK+ W+ WR C GGLG++DI+++N +LL KW+W + LW +LI KY
Sbjct: 427 REGKKIAWISWRQCCASGDVGGLGIKDIKILNNALLIKWKWLMFHQPHQLWNRILISKYK 248
Query: 235 GTRVCNLLVEDDGSWPSYISRWWKDVAFLEDEGGERWFNAEVVRKVGCGNSTSFWKDPWR 294
G R D G Y S WW D+ + + + KVG G+ FW+D W
Sbjct: 247 GWRGL-----DQGPQKYYFSPWWADLRAINQHQSMIAASNQFCWKVGRGDQILFWEDSWV 83
Query: 295 GD-IPFSRKYPRLFAISNNKETLVEE 319
D P ++P L+ +S+ + ++ +
Sbjct: 82 DDGTPLKDQFPELYRLSSQRNFIMAD 5
>TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {Arabidopsis
thaliana;} , partial (18%)
Length = 759
Score = 96.7 bits (239), Expect = 2e-20
Identities = 43/114 (37%), Positives = 69/114 (59%)
Frame = +2
Query: 131 LGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKKLCWVKWRVVC 190
+GGR+ L+ +V++++PI LSF K+P ++ K+V +Q FLWGG + ++ WVKW +C
Sbjct: 2 MGGRVTLIKSVLSALPIXXLSFFKIPQRIVDKLVTLQRQFLWGGTQHHNRIPWVKWADIC 181
Query: 191 QPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVCNLLVE 244
P+ GGLG++D+ N +L +W W + LW L E+ RV +VE
Sbjct: 182 NPKIDGGLGIKDLSNFNAALRGRWIWGLASNHNQLWAR-LAEQ*S*RVAFFVVE 340
>BI427218
Length = 421
Score = 89.7 bits (221), Expect = 2e-18
Identities = 46/143 (32%), Positives = 71/143 (49%), Gaps = 2/143 (1%)
Frame = +1
Query: 213 KWRWRVLQGEEGLWKEVLIEKYGTRVCNLLVEDDGSWPSYISRWWKDVAFLEDEGGE-RW 271
KW+W + LW +L KYG + ++ + + S WWKD+ ++ G +
Sbjct: 4 KWKWSFFYNKGELWARILDSKYG----DWRHLEEPTRGNRASAWWKDINIIDPSGADGSL 171
Query: 272 FNAEVVRKVGCGNSTSFWKDPWRGD-IPFSRKYPRLFAISNNKETLVEECRQMNGVGGGW 330
F+ + KV CG T FW+D W+ D + F KYP LF+IS + L++ G W
Sbjct: 172 FDKGIKWKVACGEKTKFWEDGWKEDRVTFKLKYPTLFSISKQ*QHLIKMMGNFTATGWEW 351
Query: 331 IFEWHRPLFVWEEELLISLKEDL 353
F+W R LF E E+ + +DL
Sbjct: 352 NFKWRRQLFDDEMEMAVKFLQDL 420
>TC231922
Length = 803
Score = 71.6 bits (174), Expect = 6e-13
Identities = 42/166 (25%), Positives = 72/166 (43%), Gaps = 9/166 (5%)
Frame = -3
Query: 254 SRWWKDVAFLEDEGGERWFNAEVVRKVGCGNSTSFWKDPWRGD-IPFSRKYPRLFAISNN 312
S WWKD+ L ++ + +V +VGCG+ FW+D W + +KY +L+ IS
Sbjct: 777 SHWWKDLRRLYNQPDFHSIHQNMVWRVGCGDKIKFWQDSWLSEGCNLQQKYNQLYTISRQ 598
Query: 313 KETLVEECRQMNGVGGGWIFEWHRPLFVWEEELLISLKEDLEG---HRWVNDPDRWVKSC 369
+ + + + W F+W R LF +E + + ++ G +D W
Sbjct: 597 *NLTISKMGKFSQNARSWEFKWRRRLFDYEYAMAVDFMNEISGISIQNQAHDSMFWKADS 418
Query: 370 YG-----KLERLLGGEVEWSLEELRVLESIWNSKAPLKVIAFSWKL 410
G RLL + R+ + +W+ K P + FSW+L
Sbjct: 417 SGVYSTKSAYRLLMPSIS-PAPSRRIFQILWHLKIPPRAAVFSWRL 283
>TC212055
Length = 776
Score = 71.2 bits (173), Expect = 8e-13
Identities = 39/108 (36%), Positives = 62/108 (57%), Gaps = 1/108 (0%)
Frame = +1
Query: 1 MRNAVSRNLFKGFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKV 60
MR A+ ++L+ + + ++ LQYADDT+ +GEA++ N+ T+K++LR FE+ASGLK+
Sbjct: 37 MREALDKSLYSSLMVGKNKIPVNILQYADDTIFLGEATMQNVMTIKSMLRVFELASGLKI 216
Query: 61 NFYKSCLMGINVPSEFMTMA-CDFLNCSEGVVPFKYLGLPVGANSFKL 107
+F KS I P + A + NC +V + L SF L
Sbjct: 217 HFAKSSFGAIGKPDHWQKEAGLNTFNCRNLIVHIRQLDQAFDKMSFIL 360
>TC221865
Length = 1045
Score = 64.7 bits (156), Expect = 7e-11
Identities = 45/145 (31%), Positives = 65/145 (44%), Gaps = 9/145 (6%)
Frame = -3
Query: 275 EVVRKVGCGNSTSFWKDPWRGD-IPFSRKYPRLFAISNNKETLVEECRQMNGVGGGWIFE 333
E+V KVGCG+ FW+D G+ RKYPR++ IS ++ L+++ W F+
Sbjct: 938 EIVWKVGCGDKFRFWEDKLVGEGETLMRKYPRMYQISCQQQQLIQQVGSHTETTWEWKFQ 759
Query: 334 WHRPLFVWEEELLISLKEDLEG---HRWVNDPDRWVKSCYGKLE-----RLLGGEVEWSL 385
W RPLF E + I ED+ HR + D W G L+ E +
Sbjct: 758 WRRPLFDNEVDTTIGFLEDISRFPIHRHLTDCWVWKSEPNGHYSTRSAYHLMQHEAAKAN 579
Query: 386 EELRVLESIWNSKAPLKVIAFSWKL 410
+ E IW K P K F+ +L
Sbjct: 578 TD-PAFEEIWKLKIPAKAAIFA*RL 507
>TC222557
Length = 1002
Score = 54.7 bits (130), Expect(2) = 1e-10
Identities = 34/101 (33%), Positives = 47/101 (45%), Gaps = 2/101 (1%)
Frame = +3
Query: 272 FNAEVVRKVGCGNSTSFWKDPW-RGDIPFSRKYPRLFAISNNKETLVEECRQMNGVGGGW 330
F + + KVGCG+ FW+D W P KYPRL+ IS ++ L+ + W
Sbjct: 51 FQSAIDWKVGCGDKIRFWEDCWLMEQEPLRAKYPRLYNISCQQQKLILVMGFHSANVWEW 230
Query: 331 IFEWHRPLFVWEEELLISLKEDLE-GHRWVNDPDRWVKSCY 370
EW R LF E + S +D+ GH DRW C+
Sbjct: 231 KLEWRRHLFDNEVQAAASFLDDISWGH-----IDRWTLDCW 338
Score = 28.9 bits (63), Expect(2) = 1e-10
Identities = 10/20 (50%), Positives = 13/20 (65%)
Frame = +1
Query: 391 LESIWNSKAPLKVIAFSWKL 410
LE +W K PLK F+W+L
Sbjct: 430 LEDLWQLKIPLKATTFAWRL 489
>BU762692
Length = 423
Score = 59.7 bits (143), Expect = 2e-09
Identities = 27/56 (48%), Positives = 35/56 (62%)
Frame = -3
Query: 48 LLRGFEMASGLKVNFYKSCLMGINVPSEFMTMACDFLNCSEGVVPFKYLGLPVGAN 103
+LR FE+ SGLK+NF KSC + + T A LNCS +PF Y G+P+GAN
Sbjct: 181 ILRTFELVSGLKINFAKSCFGAFGMTDRWKTEAASCLNCSLLAIPFVYQGIPIGAN 14
>TC213061
Length = 823
Score = 55.8 bits (133), Expect = 3e-08
Identities = 32/101 (31%), Positives = 53/101 (51%), Gaps = 12/101 (11%)
Frame = +2
Query: 327 GGGWIFEWHRPLFVWEEELLISLKEDLEGHRWVND-PDRWV-----------KSCYGKLE 374
G W F+W RPLF E E+L++ +++EGH+ +D D+WV +S Y +
Sbjct: 17 GWEWQFQWRRPLFDSEIEMLVAFIQEVEGHKIRSDIEDQWVWAAESSGSYLARSAYRVIR 196
Query: 375 RLLGGEVEWSLEELRVLESIWNSKAPLKVIAFSWKLDESLL 415
+ E E+ R + +W K P+KV F+W+L + +L
Sbjct: 197 EGIPEE-----EQDREFKELWKLKVPMKVTMFAWRLLKDIL 304
>BE554941
Length = 383
Score = 55.1 bits (131), Expect = 6e-08
Identities = 32/121 (26%), Positives = 62/121 (50%), Gaps = 1/121 (0%)
Frame = +1
Query: 199 GVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVCNLLVEDDGSWPSYISRWWK 258
G++++ + N+SLL K R L + +W +L YG+ L + + GS S S+WW+
Sbjct: 7 GIKNLSIFNISLLVKRR*HFLVETKAIWAPILKFHYGS----LDLSNTGS-ASRTSQWWR 171
Query: 259 DVAFLED-EGGERWFNAEVVRKVGCGNSTSFWKDPWRGDIPFSRKYPRLFAISNNKETLV 317
D+ + + + W + RK+G T FW+ + P + + RL +++ +K+ V
Sbjct: 172 DILRIGNCQYQPGWLTSTFCRKIGDRKDTLFWRHRCLNETPLMQSHARLXSLAMDKDISV 351
Query: 318 E 318
+
Sbjct: 352 Q 354
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 53.1 bits (126), Expect = 2e-07
Identities = 31/129 (24%), Positives = 65/129 (50%)
Frame = -2
Query: 23 SHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFYKSCLMGINVPSEFMTMACD 82
SH+ +ADD + + + N+ L + ++ + G ++ +KS + ++P + + + D
Sbjct: 451 SHVMHADDIMIFCKGTSRNVHNLINFFQLYKDSFGQVISKHKSKIFIGSIPGQRLRVISD 272
Query: 83 FLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRKLHSWGNKYVSLGGRIVLLNAVI 142
L +PF Y G+P+ + +++ KL SW +S+ GRI L+N+VI
Sbjct: 271 LLQYFHSDIPFNYFGVPIFKGKPTRRHLYCVADKIKCKLASWKGSILSIMGRIQLVNSVI 92
Query: 143 NSIPIFYLS 151
+ + ++ S
Sbjct: 91 HGMLLYSFS 65
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like
protein {Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 37.4 bits (85), Expect(2) = 6e-07
Identities = 19/61 (31%), Positives = 33/61 (53%)
Frame = -3
Query: 5 VSRNLFKGFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFYK 64
+++ L + ++ G ISHL + DD + EA++D + + L F +SG KVN K
Sbjct: 518 MNKKL*RSIQVNRGGLLISHLAFVDDLILFVEANMDQVDVINLTLELFSDSSGAKVNVDK 339
Query: 65 S 65
+
Sbjct: 338 T 336
Score = 33.9 bits (76), Expect(2) = 6e-07
Identities = 14/59 (23%), Positives = 35/59 (58%)
Frame = -2
Query: 94 KYLGLPVGANSFKLVTWEPLLEQLSRKLHSWGNKYVSLGGRIVLLNAVINSIPIFYLSF 152
KYLG+P+ + T++ ++++++++ W + +S+ GR+ L V+ ++P + F
Sbjct: 246 KYLGVPIFHKNPTSHTFDFIIDKVNKRFSRWKSNLLSMVGRLTLTK*VV*ALPSHIMQF 70
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.141 0.472
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,185,809
Number of Sequences: 63676
Number of extensions: 479319
Number of successful extensions: 2205
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 2168
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2181
length of query: 420
length of database: 12,639,632
effective HSP length: 100
effective length of query: 320
effective length of database: 6,272,032
effective search space: 2007050240
effective search space used: 2007050240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC122162.14


