

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122160.6 + phase: 0
(466 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
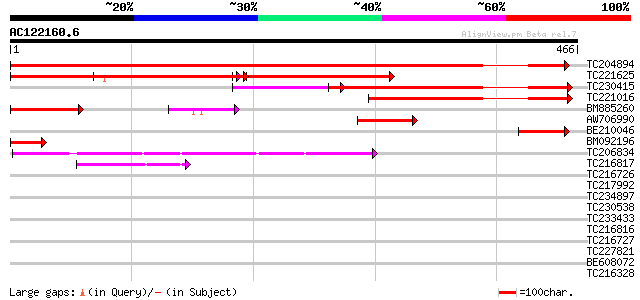
Score E
Sequences producing significant alignments: (bits) Value
TC204894 similar to UP|Q94KA9 (Q94KA9) Importin alpha 2, partial... 634 0.0
TC221625 similar to UP|O82783 (O82783) Importin alpha 2 subunit ... 292 e-120
TC230415 similar to GB|AAB72116.2|13699777|ATU69533 AtKAP alpha ... 255 3e-68
TC221016 similar to GB|AAB72116.2|13699777|ATU69533 AtKAP alpha ... 186 2e-47
BM885260 similar to GP|16974501|gb AT3g06720/F3E22_14 {Arabidops... 84 1e-16
AW706990 similar to GP|13699777|gb| AtKAP alpha {Arabidopsis tha... 81 9e-16
BE210046 similar to GP|13752562|gb| importin alpha 2 {Capsicum a... 70 2e-12
BM092196 similar to GP|13752562|gb importin alpha 2 {Capsicum an... 49 6e-06
TC206834 similar to UP|Q944I3 (Q944I3) AT3g01400/T13O15_4, parti... 47 1e-05
TC216817 weakly similar to UP|KHLX_HUMAN (Q9Y2M5) Kelch-like pro... 45 7e-05
TC216726 similar to UP|Q9SNC6 (Q9SNC6) Arm repeat containing pro... 40 0.003
TC217992 38 0.008
TC234897 weakly similar to UP|TIR3_YEAST (P40552) Cell wall prot... 38 0.008
TC230538 34 0.16
TC233433 33 0.21
TC216816 weakly similar to UP|KHLX_HUMAN (Q9Y2M5) Kelch-like pro... 33 0.27
TC216727 similar to UP|Q9ZV31 (Q9ZV31) Expressed protein, partia... 33 0.35
TC227821 weakly similar to UP|Q7PY15 (Q7PY15) EbiP8519 (Fragment... 32 0.46
BE608072 weakly similar to GP|22830929|db P0466H10.27 {Oryza sat... 32 0.46
TC216328 weakly similar to UP|Q6L3M1 (Q6L3M1) Expressed protein,... 30 1.8
>TC204894 similar to UP|Q94KA9 (Q94KA9) Importin alpha 2, partial (95%)
Length = 2332
Score = 634 bits (1636), Expect = 0.0
Identities = 326/462 (70%), Positives = 371/462 (79%), Gaps = 2/462 (0%)
Frame = +2
Query: 1 MVADIWSDDNSQQLEATTEFRKRLSVDK-PPIDDVIQSGVVPRFVQFLDKGDFPQLQLEA 59
MV +W+DDN+ QLEATT+FRK LS+++ PPI++VIQ+GVV RFV+FL + DFPQLQ EA
Sbjct: 395 MVTGVWTDDNNLQLEATTQFRKLLSIERSPPIEEVIQTGVVSRFVEFLMREDFPQLQFEA 574
Query: 60 AWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLV 119
AWALTNIA+GTSENTKVV+DHGAVP+FVKLL+SP DDVR QA WALGN+AGDSPR RDLV
Sbjct: 575 AWALTNIASGTSENTKVVIDHGAVPIFVKLLASPSDDVREQAVWALGNVAGDSPRCRDLV 754
Query: 120 LSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDE 179
LSHGAL+ LL+QLNE L LRNA WTLSNFCRGKPQP +QV+ ALPAL L+ S DE
Sbjct: 755 LSHGALLPLLAQLNEHAKLSMLRNATWTLSNFCRGKPQPPFDQVKPALPALARLIHSNDE 934
Query: 180 VVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDD 239
VLT+ACWALSYLSDGT+D IQAVIEAGVC RLV++LL PS S L PALRTVGNIV+GDD
Sbjct: 935 EVLTDACWALSYLSDGTNDKIQAVIEAGVCPRLVELLLHPSPSVLIPALRTVGNIVTGDD 1114
Query: 240 MQTQAIVNHGLLPCLLSLLTHP-KKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVN 298
MQTQ I+NH LPCLL+LLT+ KKSIKKEAC T+SNITAGN++QIQAVIEA LIAPLVN
Sbjct: 1115MQTQVIINHQALPCLLNLLTNNYKKSIKKEACWTISNITAGNKQQIQAVIEANLIAPLVN 1294
Query: 299 LLQNAEFDITKEVAQALTNVTSGGTHEHIKYLVSQGCIKPLCDLLVCPDPRIVTVCLEGL 358
LLQNAEFDI KE A A++N TSGG+HE IK+LVSQGCIKPLCDLL+CPDPRIVTVCLEGL
Sbjct: 1295LLQNAEFDIKKEAAWAISNATSGGSHEQIKFLVSQGCIKPLCDLLICPDPRIVTVCLEGL 1474
Query: 359 ENFLKVGEAEKSISNTGDVNLYAQTIKDVEGLEKGLEKFLKVGEAEKRFGNTGDVNLYAQ 418
EN LKVGEA+K+I NTGDVNLYAQ I + EG
Sbjct: 1475ENILKVGEADKNIGNTGDVNLYAQMIDEAEG----------------------------- 1567
Query: 419 MIEDVKGLEKIENLDSHEDREIYEKAAKILERYWLEDEDETL 460
LEKIENL SH++ EIYEKA KILE YWLE+EDET+
Sbjct: 1568-------LEKIENLQSHDNTEIYEKAVKILETYWLEEEDETM 1672
>TC221625 similar to UP|O82783 (O82783) Importin alpha 2 subunit (NLS
receptor), partial (62%)
Length = 988
Score = 292 bits (747), Expect(2) = e-120
Identities = 144/197 (73%), Positives = 167/197 (84%), Gaps = 1/197 (0%)
Frame = +3
Query: 1 MVADIWSDDNSQQLEATTEFRKRLSVDK-PPIDDVIQSGVVPRFVQFLDKGDFPQLQLEA 59
MV +++DDN+ QLEATT+FRK LS+++ PPI++VIQ+GVVPRFV FL + DFPQLQ EA
Sbjct: 33 MVTGVFTDDNNMQLEATTQFRKLLSIERSPPIEEVIQAGVVPRFVDFLMREDFPQLQFEA 212
Query: 60 AWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLV 119
AWALTNIA+GTSENTKV++DHGAVP+FVKLL SP DDVR QA WALGN+AGDSPR RDLV
Sbjct: 213 AWALTNIASGTSENTKVIIDHGAVPIFVKLLGSPSDDVREQAVWALGNVAGDSPRCRDLV 392
Query: 120 LSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDE 179
L HGAL+ LL+QLNE L LRNA WTLSNFCRGKPQPA +QV+ ALPAL L+ S DE
Sbjct: 393 LGHGALLPLLAQLNEHAKLSMLRNATWTLSNFCRGKPQPAFDQVKPALPALASLIQSTDE 572
Query: 180 VVLTEACWALSYLSDGT 196
VLT+ACWALSYLSD T
Sbjct: 573 EVLTDACWALSYLSDRT 623
Score = 160 bits (404), Expect(2) = e-120
Identities = 85/123 (69%), Positives = 101/123 (82%), Gaps = 1/123 (0%)
Frame = +1
Query: 195 GTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDMQTQAIVNHGLLPCL 254
G +D IQ VIEAGVC RL ++LL PS S L PALRTVGNIV+GDD+QT+ I+NH L L
Sbjct: 619 GHNDKIQGVIEAGVCSRLGELLLHPSPSVLIPALRTVGNIVTGDDLQTEVIINHQALSRL 798
Query: 255 LSLLTHP-KKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQNAEFDITKEVAQ 313
L+LLT+ KKSIKKEAC T+SNITAGN++QIQ VIEA +IAPLV+LLQNAEFDI KE A
Sbjct: 799 LNLLTNNYKKSIKKEACWTISNITAGNKKQIQDVIEASIIAPLVHLLQNAEFDIKKEAAW 978
Query: 314 ALT 316
A++
Sbjct: 979 AIS 987
Score = 63.9 bits (154), Expect = 1e-10
Identities = 41/134 (30%), Positives = 68/134 (50%), Gaps = 1/134 (0%)
Frame = +3
Query: 184 EACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDMQTQ 243
EA WAL+ ++ GTS+N + +I+ G V++L PS A+ +GN+
Sbjct: 207 EAAWALTNIASGTSENTKVIIDHGAVPIFVKLLGSPSDDVREQAVWALGNVAGDSPRCRD 386
Query: 244 AIVNHG-LLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQN 302
++ HG LLP L L H K S+ + A TLSN G + ++ L A L +L+Q+
Sbjct: 387 LVLGHGALLPLLAQLNEHAKLSMLRNATWTLSNFCRGKPQPAFDQVKPALPA-LASLIQS 563
Query: 303 AEFDITKEVAQALT 316
+ ++ + AL+
Sbjct: 564 TDEEVLTDACWALS 605
Score = 43.5 bits (101), Expect = 2e-04
Identities = 33/124 (26%), Positives = 57/124 (45%), Gaps = 3/124 (2%)
Frame = +1
Query: 70 TSENTKV--VVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVLSHGALIL 127
T N K+ V++ G +LL P V A +GNI +++++H AL
Sbjct: 616 TGHNDKIQGVIEAGVCSRLGELLLHPSPSVLIPALRTVGNIVTGDDLQTEVIINHQALSR 795
Query: 128 LLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPA-LKCLVFSKDEVVLTEAC 186
LL+ L + A WT+SN G + + + +++ A L L+ + + + EA
Sbjct: 796 LLNLLTNNYKKSIKKEACWTISNITAGNKKQIQDVIEASIIAPLVHLLQNAEFDIKKEAA 975
Query: 187 WALS 190
WA+S
Sbjct: 976 WAIS 987
>TC230415 similar to GB|AAB72116.2|13699777|ATU69533 AtKAP alpha {Arabidopsis
thaliana;} , partial (34%)
Length = 971
Score = 255 bits (651), Expect = 3e-68
Identities = 135/200 (67%), Positives = 144/200 (71%)
Frame = +1
Query: 263 KSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQNAEFDITKEVAQALTNVTSGG 322
KSIKKEAC T+SNITAGN+EQIQ VIEAGL+APLVNLLQNAEFDI KE A A++N TSGG
Sbjct: 1 KSIKKEACWTISNITAGNKEQIQTVIEAGLVAPLVNLLQNAEFDIKKEAAWAISNATSGG 180
Query: 323 THEHIKYLVSQGCIKPLCDLLVCPDPRIVTVCLEGLENFLKVGEAEKSISNTGDVNLYAQ 382
HE IKYLVSQGCIKPLCDLLVCPDPRIVTVCLEGLEN LKVGEAEKS+ NTGDVN YAQ
Sbjct: 181 IHEQIKYLVSQGCIKPLCDLLVCPDPRIVTVCLEGLENILKVGEAEKSLGNTGDVNEYAQ 360
Query: 383 TIKDVEGLEKGLEKFLKVGEAEKRFGNTGDVNLYAQMIEDVKGLEKIENLDSHEDREIYE 442
I D EG LEKIENL H++ EIYE
Sbjct: 361 MIDDAEG------------------------------------LEKIENLQRHDNNEIYE 432
Query: 443 KAAKILERYWLEDEDETLTS 462
KA KILE YWLE+EDETL S
Sbjct: 433 KAVKILETYWLEEEDETLPS 492
Score = 48.1 bits (113), Expect = 8e-06
Identities = 31/94 (32%), Positives = 42/94 (43%), Gaps = 1/94 (1%)
Frame = +1
Query: 184 EACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGD-DMQT 242
EACW +S ++ G + IQ VIEAG+ LV +L A + N SG Q
Sbjct: 16 EACWTISNITAGNKEQIQTVIEAGLVAPLVNLLQNAEFDIKKEAAWAISNATSGGIHEQI 195
Query: 243 QAIVNHGLLPCLLSLLTHPKKSIKKEACRTLSNI 276
+ +V+ G + L LL P I L NI
Sbjct: 196 KYLVSQGCIKPLCDLLVCPDPRIVTVCLEGLENI 297
Score = 39.7 bits (91), Expect = 0.003
Identities = 32/118 (27%), Positives = 53/118 (44%), Gaps = 1/118 (0%)
Frame = +1
Query: 27 DKPPIDDVIQSGVVPRFVQFLDKGDFPQLQLEAAWALTN-IAAGTSENTKVVVDHGAVPM 85
+K I VI++G+V V L +F ++ EAAWA++N + G E K +V G +
Sbjct: 52 NKEQIQTVIEAGLVAPLVNLLQNAEF-DIKKEAAWAISNATSGGIHEQIKYLVSQGCIKP 228
Query: 86 FVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVLSHGALILLLSQLNEQEGLYRLRN 143
LL P + L NI + L + G + +++ EGL ++ N
Sbjct: 229 LCDLLVCPDPRIVTVCLEGLENILKVGEAEKSLG-NTGDVNEYAQMIDDAEGLEKIEN 399
>TC221016 similar to GB|AAB72116.2|13699777|ATU69533 AtKAP alpha {Arabidopsis
thaliana;} , partial (27%)
Length = 851
Score = 186 bits (472), Expect = 2e-47
Identities = 98/167 (58%), Positives = 109/167 (64%)
Frame = +1
Query: 296 LVNLLQNAEFDITKEVAQALTNVTSGGTHEHIKYLVSQGCIKPLCDLLVCPDPRIVTVCL 355
L NLL AEFD E + A++N TSGGTH+ IKYLVSQGC+KPLCDLLVCPD RIVTVCL
Sbjct: 1 LXNLLXXAEFDXXXEASWAISNATSGGTHDQIKYLVSQGCVKPLCDLLVCPDXRIVTVCL 180
Query: 356 EGLENFLKVGEAEKSISNTGDVNLYAQTIKDVEGLEKGLEKFLKVGEAEKRFGNTGDVNL 415
EGLEN LKVGEAEKS+ NTGDVN+YAQ I + EG
Sbjct: 181 EGLENILKVGEAEKSLGNTGDVNVYAQMIDEAEG-------------------------- 282
Query: 416 YAQMIEDVKGLEKIENLDSHEDREIYEKAAKILERYWLEDEDETLTS 462
LEKIENL SH++ EIYEK KILE YWLE+EDETL S
Sbjct: 283 ----------LEKIENLQSHDNNEIYEKVVKILETYWLEEEDETLPS 393
>BM885260 similar to GP|16974501|gb AT3g06720/F3E22_14 {Arabidopsis
thaliana}, partial (25%)
Length = 423
Score = 84.0 bits (206), Expect = 1e-16
Identities = 40/61 (65%), Positives = 52/61 (84%), Gaps = 1/61 (1%)
Frame = +1
Query: 1 MVADIWSDDNSQQLEATTEFRKRLSVDK-PPIDDVIQSGVVPRFVQFLDKGDFPQLQLEA 59
MV +W+DDN+ QLEATT+FRK LS+++ PPI++VIQ+GVV RFV+FL + DFPQLQ EA
Sbjct: 145 MVTGVWTDDNNLQLEATTQFRKLLSIERSPPIEEVIQTGVVSRFVEFLMREDFPQLQFEA 324
Query: 60 A 60
A
Sbjct: 325 A 327
Score = 50.4 bits (119), Expect = 2e-06
Identities = 32/68 (47%), Positives = 38/68 (55%), Gaps = 9/68 (13%)
Frame = +2
Query: 131 QLNEQEGLYRLRNAVWTLS--NFCRGKP-------QPALEQVRSALPALKCLVFSKDEVV 181
+LN L RL VW L +F GK QP +QV+ ALPAL L+ S DE V
Sbjct: 218 RLNVVHRLRRLYRLVWFLDLLSFS*GKTFRSCSLRQPPFDQVKPALPALARLIHSNDEEV 397
Query: 182 LTEACWAL 189
LT+ACWAL
Sbjct: 398 LTDACWAL 421
>AW706990 similar to GP|13699777|gb| AtKAP alpha {Arabidopsis thaliana},
partial (9%)
Length = 419
Score = 81.3 bits (199), Expect = 9e-16
Identities = 38/49 (77%), Positives = 44/49 (89%)
Frame = +1
Query: 287 VIEAGLIAPLVNLLQNAEFDITKEVAQALTNVTSGGTHEHIKYLVSQGC 335
VIEAGL+APLVNLLQ+AEFDI KE + A++N TSGGTH+ IKYLVSQGC
Sbjct: 271 VIEAGLVAPLVNLLQSAEFDIKKEASWAISNATSGGTHDQIKYLVSQGC 417
>BE210046 similar to GP|13752562|gb| importin alpha 2 {Capsicum annuum},
partial (12%)
Length = 362
Score = 70.1 bits (170), Expect = 2e-12
Identities = 31/42 (73%), Positives = 37/42 (87%)
Frame = -1
Query: 419 MIEDVKGLEKIENLDSHEDREIYEKAAKILERYWLEDEDETL 460
MI++ +GLEKIENL SH++ EIYEKA KILE YWLED+DETL
Sbjct: 362 MIDEAEGLEKIENLQSHDNNEIYEKAVKILETYWLEDDDETL 237
>BM092196 similar to GP|13752562|gb importin alpha 2 {Capsicum annuum},
partial (20%)
Length = 375
Score = 48.5 bits (114), Expect = 6e-06
Identities = 21/30 (70%), Positives = 26/30 (86%)
Frame = +1
Query: 1 MVADIWSDDNSQQLEATTEFRKRLSVDKPP 30
MVA +WSDDNS QLEATT+FRK LS+++ P
Sbjct: 286 MVAGVWSDDNSIQLEATTQFRKLLSIERSP 375
>TC206834 similar to UP|Q944I3 (Q944I3) AT3g01400/T13O15_4, partial (89%)
Length = 1740
Score = 47.4 bits (111), Expect = 1e-05
Identities = 68/301 (22%), Positives = 129/301 (42%), Gaps = 1/301 (0%)
Frame = +3
Query: 3 ADIWSDDNSQQLEATTEFRKRL-SVDKPPIDDVIQSGVVPRFVQFLDKGDFPQLQLEAAW 61
+D SD + + A+++ R+ L + DD+I+ V +D Q +AA
Sbjct: 132 SDCNSDRSGEFATASSQTRRFLIACATENSDDLIRQLVADLHSSSIDD------QKQAAM 293
Query: 62 ALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVLS 121
+ +A EN + GA+ + L+SSP ++ A+ N++ ++++ S
Sbjct: 294 EIRLLAKNKPENRIKIAKAGAIKPLISLISSPDLQLQEYGVTAILNLS-LCDENKEVIAS 470
Query: 122 HGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDEVV 181
GA+ L+ LN + NA L + + A A+P L L+ S
Sbjct: 471 SGAIKPLVRALNSGTATAK-ENAACALLRLSQVEENKAAIGRSGAIPLLVSLLESGGFRA 647
Query: 182 LTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDMQ 241
+A AL L + I+AV +AG+ LV+++ + + + + V +V+ + +
Sbjct: 648 KKDASTALYSLCTVKENKIRAV-KAGIMKVLVELMADFESNMVDKSAYVVSVLVAVPEAR 824
Query: 242 TQAIVNHGLLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQ 301
A+V G +P L+ ++ + K+ L + + V G I PLV+L Q
Sbjct: 825 V-ALVEEGGVPVLVEIVEVGTQRQKEIVVVILLQVCEDSVAYRTMVAREGAIPPLVSLSQ 1001
Query: 302 N 302
+
Sbjct: 1002S 1004
>TC216817 weakly similar to UP|KHLX_HUMAN (Q9Y2M5) Kelch-like protein X,
partial (5%)
Length = 1071
Score = 45.1 bits (105), Expect = 7e-05
Identities = 32/93 (34%), Positives = 49/93 (52%)
Frame = +1
Query: 56 QLEAAWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRG 115
Q EAA L AA S+ +V GAV +++L SP +R +A+ALG +A D P
Sbjct: 175 QREAALLLGQFAATDSDCKVHIVQRGAVQPLIEMLQSPDVQLREMSAFALGRLAQD-PHN 351
Query: 116 RDLVLSHGALILLLSQLNEQEGLYRLRNAVWTL 148
+ + +G L+ LL L+ + G + NA + L
Sbjct: 352 QAGIAHNGGLVPLLKLLDSKNGSLQ-HNAAFAL 447
Score = 38.5 bits (88), Expect = 0.006
Identities = 65/291 (22%), Positives = 101/291 (34%), Gaps = 13/291 (4%)
Frame = +1
Query: 82 AVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVLSHGALILLLSQLNEQEGLYRL 141
A+P + +L S V +A +GN+ SP + VL GA L
Sbjct: 1 ALPTLILMLRSEDAGVHYEAVGVIGNLVHSSPNIKKEVLLAGA----------------L 132
Query: 142 RNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDEVVLTEACWALSYLSDGTSDNIQ 201
+ + LS+ C + EA L + SD
Sbjct: 133 QPVIGLLSSCCSESQR--------------------------EAALLLGQFAATDSDCKV 234
Query: 202 AVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDMQTQAIVNHGLLPCLLSLLTHP 261
+++ G L+++L P + + +G + Q N GL+P LL LL
Sbjct: 235 HIVQRGAVQPLIEMLQSPDVQLREMSAFALGRLAQDPHNQAGIAHNGGLVP-LLKLLDSK 411
Query: 262 KKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQNAEF------DITKEVAQAL 315
S++ A L + A N + I G V LQ+ EF D + + L
Sbjct: 412 NGSLQHNAAFALYGL-ADNEDNASDFIRVG----GVQRLQDGEFIVQATKDCVAKTLKRL 576
Query: 316 TNVTSGGTHEHIKYLV---SQGCIK----PLCDLLVCPDPRIVTVCLEGLE 359
G H+ YL+ +GC + L L D RI+ + GLE
Sbjct: 577 EEKIHGRVLNHLLYLMRVSEKGCQRRVALALAHLCSSDDQRIIFIDHYGLE 729
>TC216726 similar to UP|Q9SNC6 (Q9SNC6) Arm repeat containing protein
homolog, partial (35%)
Length = 1115
Score = 39.7 bits (91), Expect = 0.003
Identities = 36/154 (23%), Positives = 71/154 (45%)
Frame = +3
Query: 166 ALPALKCLVFSKDEVVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALF 225
A+P L L+ ++ +A AL L + +AV AGV L+++L EPS +
Sbjct: 216 AIPPLVTLLSEGNQRGKKDAATALFNLCIYQGNKGKAV-RAGVIPTLMRLLTEPSGGMVD 392
Query: 226 PALRTVGNIVSGDDMQTQAIVNHGLLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQIQ 285
AL + + S + + + +P L+ + + K+ A L ++ +G+++ +
Sbjct: 393 EALAILAILASHPEGKATIRASEA-VPVLVEFIGNGSPRNKENAAAVLVHLCSGDQQYLA 569
Query: 286 AVIEAGLIAPLVNLLQNAEFDITKEVAQALTNVT 319
E G++ PL+ L QN ++ Q L ++
Sbjct: 570 QAQELGVMGPLLELAQNGTDRGKRKAGQLLERMS 671
Score = 37.7 bits (86), Expect = 0.011
Identities = 52/208 (25%), Positives = 83/208 (39%), Gaps = 1/208 (0%)
Frame = +3
Query: 9 DNSQQLEATTEFRKRLSVDKPPIDDVIQSGVVPRFVQFLDKGDFPQLQLEAAWALTNIAA 68
D+ Q A T LS+ + ++ SG VP V L KG + + AA L +++
Sbjct: 6 DSRTQEHAVTALLN-LSIYENNKGSIVSSGAVPGIVHVLKKGSM-EARENAAATLFSLSV 179
Query: 69 GTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIA-GDSPRGRDLVLSHGALIL 127
EN + GA+P V LLS + AA AL N+ +G+ + G +
Sbjct: 180 -IDENKVTIGSLGAIPPLVTLLSEGNQRGKKDAATALFNLCIYQGNKGK--AVRAGVIPT 350
Query: 128 LLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDEVVLTEACW 187
L+ L E G + A+ L+ A + A+P L + + A
Sbjct: 351 LMRLLTEPSG-GMVDEALAILAILASHPEGKATIRASEAVPVLVEFIGNGSPRNKENAAA 527
Query: 188 ALSYLSDGTSDNIQAVIEAGVCGRLVQI 215
L +L G + E GV G L+++
Sbjct: 528 VLVHLCSGDQQYLAQAQELGVMGPLLEL 611
>TC217992
Length = 1097
Score = 38.1 bits (87), Expect = 0.008
Identities = 34/108 (31%), Positives = 56/108 (51%), Gaps = 1/108 (0%)
Frame = +1
Query: 243 QAIVNHGLLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQN 302
+AIV HG + L++L + A TL NI+A E QA+ E G++ ++NLL
Sbjct: 739 RAIVGHGGVRPLVALCQTGDSVSQAAAACTLKNISA-VPEVRQALAEEGIVTVMINLLNC 915
Query: 303 AEFDITKE-VAQALTNVTSGGTHEHIKYLVSQGCIKPLCDLLVCPDPR 349
+KE A+ L N+T+ + K ++S+G ++ L L P P+
Sbjct: 916 GILLGSKEHAAECLQNLTASNENLR-KSVISEGGVRSLLAYLDGPLPQ 1056
>TC234897 weakly similar to UP|TIR3_YEAST (P40552) Cell wall protein TIR3
precursor, partial (12%)
Length = 698
Score = 38.1 bits (87), Expect = 0.008
Identities = 37/109 (33%), Positives = 56/109 (50%), Gaps = 7/109 (6%)
Frame = +3
Query: 29 PPI--DDVIQSGVVPRFVQFLDKGDFPQLQLEAAWALTNIAAGTSENTKVVVDHGAVPMF 86
PPI +D I S V F+ + G ++EAA L + A + N K++V+ VP
Sbjct: 345 PPIASNDPILSWVWS-FIASIQMGQLND-RIEAANELASFAQDNARNKKIIVEECGVPPL 518
Query: 87 VKLL---SSPCDDVRGQAAWALGNIAGDSPRGRDLVLSHG--ALILLLS 130
+KLL +SP + AA L ++A D R R +V HG A++ +LS
Sbjct: 519 LKLLKEGTSPLAQI--AAATTLCHLANDLDRVRVIVSEHGVPAVVQVLS 659
>TC230538
Length = 1261
Score = 33.9 bits (76), Expect = 0.16
Identities = 45/188 (23%), Positives = 78/188 (40%), Gaps = 8/188 (4%)
Frame = +2
Query: 187 WALSYLSDGTSDNIQAVIEAGVCGRLV-QILLEPSL--SALFPAL--RTVGNIVSGDDMQ 241
+ALS LS+ + + + LV QI E SL +P L R ++ +
Sbjct: 416 FALSSLSEQLLETQETGFDTSSLKHLVEQIFTEDSLIDPLEYPFLYARIFTSVAKLSSLI 595
Query: 242 TQAIVNHGLLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQ 301
+ ++ H L + ++ +K ACR L+N+ +++I GLI+ L +LL
Sbjct: 596 SNGLLEHFLYLAMKAITMDVPPPVKVGACRALTNLLPEAKKEIVQSQLLGLISSLTDLLN 775
Query: 302 NAEFDITKEVAQALTNVTSGGTHEH---IKYLVSQGCIKPLCDLLVCPDPRIVTVCLEGL 358
+A + V L G HE +++++S + + DP I LE L
Sbjct: 776 HASDETLLMVLDTLLAAVKAG-HESSTLVEHMISPVILNVWASHV--SDPFISIDALEVL 946
Query: 359 ENFLKVGE 366
E + E
Sbjct: 947 EAIKSIPE 970
>TC233433
Length = 754
Score = 33.5 bits (75), Expect = 0.21
Identities = 23/70 (32%), Positives = 34/70 (47%), Gaps = 3/70 (4%)
Frame = +2
Query: 277 TAGNREQIQAVIEAGLIAPLVNLLQNAEFDITKEVAQALTN--VTSGGTH-EHIKYLVSQ 333
T GN + E +I PLV LL E ++++E + ALT T H +H K ++
Sbjct: 458 TIGNLARTFKATETRMIGPLVKLLDEREAEVSREASIALTKFACTENYLHVDHSKAIIIA 637
Query: 334 GCIKPLCDLL 343
G K L L+
Sbjct: 638 GGAKHLIQLV 667
>TC216816 weakly similar to UP|KHLX_HUMAN (Q9Y2M5) Kelch-like protein X,
partial (5%)
Length = 1610
Score = 33.1 bits (74), Expect = 0.27
Identities = 25/82 (30%), Positives = 44/82 (53%), Gaps = 1/82 (1%)
Frame = +1
Query: 68 AGTSENTKV-VVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVLSHGALI 126
A T + KV +V GAV +++L S ++ +A+ALG +A D+ + + +G L+
Sbjct: 28 AATDSDCKVHIVQRGAVRPLIEMLQSSDVQLKEMSAFALGRLAQDT-HNQAGIAHNGGLM 204
Query: 127 LLLSQLNEQEGLYRLRNAVWTL 148
LL L+ + G + NA + L
Sbjct: 205 PLLKLLDSKNGSLQ-HNAAFAL 267
>TC216727 similar to UP|Q9ZV31 (Q9ZV31) Expressed protein, partial (28%)
Length = 654
Score = 32.7 bits (73), Expect = 0.35
Identities = 18/57 (31%), Positives = 30/57 (52%)
Frame = +2
Query: 53 PQLQLEAAWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIA 109
P+ Q AA + +A ++N + + GA+P+ V LLS P + A AL N++
Sbjct: 269 PEDQRSAAGEIRLLAKRNADNRVAIAEAGAIPLLVGLLSVPDSRTQEHAVTALLNLS 439
Score = 28.1 bits (61), Expect = 8.7
Identities = 35/152 (23%), Positives = 63/152 (41%), Gaps = 13/152 (8%)
Frame = +2
Query: 139 YRLRNAV--WTLSNFCRGKPQPALEQVRSALPALKCLVFSKDEVVLTE-----------A 185
Y LR+ + W +N +P+ Q + A SK E +L + A
Sbjct: 110 YVLRSLIAQWCEANGIEPPKRPSDSQPSKSASAYSPAEQSKIESLLQKLTSVSPEDQRSA 289
Query: 186 CWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDMQTQAI 245
+ L+ +DN A+ EAG LV +L P A+ + N+ S + +I
Sbjct: 290 AGEIRLLAKRNADNRVAIAEAGAIPLLVGLLSVPDSRTQEHAVTALLNL-SIYENNKGSI 466
Query: 246 VNHGLLPCLLSLLTHPKKSIKKEACRTLSNIT 277
V+ G +P ++ +L ++ A TL +++
Sbjct: 467 VSSGAVPGIVHVLKKGSMEARENAAATLFSLS 562
>TC227821 weakly similar to UP|Q7PY15 (Q7PY15) EbiP8519 (Fragment), partial
(5%)
Length = 1743
Score = 32.3 bits (72), Expect = 0.46
Identities = 64/264 (24%), Positives = 109/264 (41%), Gaps = 13/264 (4%)
Frame = +3
Query: 86 FVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVLSHGALILLLSQLNEQEGLYRLRNAV 145
FVK L VR AA ALG ++G S R LV LLS L +G +R+A+
Sbjct: 540 FVKCLQDSTRTVRSSAALALGKLSGLSTRVDPLVSD------LLSSLQGSDG--GVRDAI 695
Query: 146 WTLSNFCRGKPQPALEQVRSAL-----PALKCLVFSKDEVVLTEACWALSYLSDGTSDNI 200
T +G + A + + SA+ LK L+ D+ V T A L L+ D +
Sbjct: 696 LTA---LKGVLKHAGKNLSSAVRTRFYSILKDLIHDDDDRVRTYASSILGILTQYLED-V 863
Query: 201 QAVIEAGVCGRLVQILLEPSLSALFP----ALRTVGNIVSGDDMQTQAIVNHGLLP---- 252
Q L+Q L + S+ +P ++ T+ +++ I + L P
Sbjct: 864 QLT-------ELIQELSSLANSSSWPPRHGSILTISSLL---HYNPATICSSSLFPTIVD 1013
Query: 253 CLLSLLTHPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQNAEFDITKEVA 312
CL L K +++ + + L + R Q+ + L +++LL ++ D + EV
Sbjct: 1014CLRDTLKDEKFPLRETSTKALGRLLL-YRSQVDP-SDTLLYKDVLSLLVSSTHDDSSEVR 1187
Query: 313 QALTNVTSGGTHEHIKYLVSQGCI 336
+ + + ++S G I
Sbjct: 1188RRALSAIKAVAKANPSAIMSLGTI 1259
>BE608072 weakly similar to GP|22830929|db P0466H10.27 {Oryza sativa
(japonica cultivar-group)}, partial (20%)
Length = 473
Score = 32.3 bits (72), Expect = 0.46
Identities = 24/94 (25%), Positives = 44/94 (46%), Gaps = 7/94 (7%)
Frame = +3
Query: 27 DKPPIDDVIQSGVVPRFVQFLDKGDFPQ-------LQLEAAWALTNIAAGTSENTKVVVD 79
DK D + ++ ++P V +D G ++ A A+ ++ + +
Sbjct: 30 DKKVTDALKRANLLPILVSIMDLGTGSNSPAKSILMESIAGIAIRFTSSSDKKLQLLSAK 209
Query: 80 HGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSP 113
HG + + VKLLSS + +AA ALG ++ +SP
Sbjct: 210 HGVIALLVKLLSSGSAITKLKAATALGQLSQNSP 311
>TC216328 weakly similar to UP|Q6L3M1 (Q6L3M1) Expressed protein, partial
(41%)
Length = 450
Score = 30.4 bits (67), Expect = 1.8
Identities = 25/89 (28%), Positives = 46/89 (51%), Gaps = 6/89 (6%)
Frame = +1
Query: 221 LSALFPALRTVG-NIVSG-----DDMQTQAIVNHGLLPCLLSLLTHPKKSIKKEACRTLS 274
LS+ P +R +IV G + +Q+ A ++ LLP L LLT PK+ + + A L
Sbjct: 58 LSSPSPQIRKAAVDIVRGLTGSVEGLQSLANYSNALLPALSRLLTLPKE-VSEAAAEALV 234
Query: 275 NITAGNREQIQAVIEAGLIAPLVNLLQNA 303
N++ N + ++ GL+ +++ + A
Sbjct: 235 NLSQ-NSSLAEVMVRMGLVKTAMDVSEQA 318
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,145,077
Number of Sequences: 63676
Number of extensions: 242056
Number of successful extensions: 1132
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 1093
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1115
length of query: 466
length of database: 12,639,632
effective HSP length: 101
effective length of query: 365
effective length of database: 6,208,356
effective search space: 2266049940
effective search space used: 2266049940
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122160.6


