

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122160.2 - phase: 0
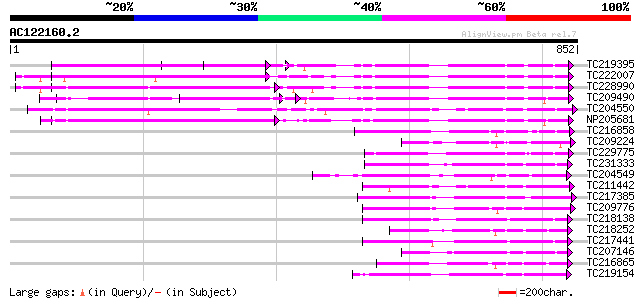
(852 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, part... 292 4e-79
TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, comp... 290 2e-78
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 287 2e-77
TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment),... 263 3e-70
TC204550 UP|Q8L3Y5 (Q8L3Y5) Receptor-like kinase RHG1, complete 262 5e-70
NP205681 receptor protein kinase-like protein 258 1e-68
TC216858 homologue to UP|Q76FZ8 (Q76FZ8) Brassinosteroid recepto... 230 2e-60
TC209224 similar to UP|BRI1_LYCPE (Q8L899) Systemin receptor SR1... 188 8e-48
TC229775 weakly similar to UP|KPEL_DROME (Q05652) Probable serin... 169 6e-42
TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, parti... 166 4e-41
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 165 7e-41
TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicite... 165 7e-41
TC217385 similar to UP|Q6ZGC7 (Q6ZGC7) Receptor protein kinase P... 164 2e-40
TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-l... 163 3e-40
TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis r... 157 1e-38
TC218252 weakly similar to UP|Q03407 (Q03407) Ydr490cp, partial ... 157 2e-38
TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic em... 155 5e-38
TC207146 weakly similar to UP|Q9H4D1 (Q9H4D1) Protein kinase, pa... 152 6e-37
TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial... 151 1e-36
TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.1... 149 7e-36
>TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, partial (92%)
Length = 3037
Score = 292 bits (748), Expect = 4e-79
Identities = 252/806 (31%), Positives = 386/806 (47%), Gaps = 22/806 (2%)
Frame = +3
Query: 64 NKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLS 122
N +V L+ + GL+G IP +GKL KL +L L N ++ +L + +L SLKS++LS
Sbjct: 468 NLSELVRLDAAYCGLSGEIP-AALGKLQKLDTLFLQVNALSGSLTPELGNLKSLKSMDLS 644
Query: 123 SNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGI 182
+N +SG + G + +L +N IPE + L +L+V++L N F SIP G+
Sbjct: 645 NNMLSGEIPARFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNFTGSIPEGL 824
Query: 183 LKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNI 241
K L +DLSSN+L+GTLP L+TL N ++G + + +S+ + +
Sbjct: 825 GKNGRLNLVDLSSNKLTGTLPTYLCSG-NTLQTLITLGNFLFGPIPESLGSCESLTRIRM 1001
Query: 242 SGNSFQGSIIE-VFVL-KLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQN 299
N GSI +F L KL ++L N G +V +L + LS NQLSG + +
Sbjct: 1002 GENFLNGSIPRGLFGLPKLTQVELQDNYLSGEFPEVGSVAVNLGQITLSNNQLSGVLPPS 1181
Query: 300 LNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDL 359
+ N +++ L L N F+ + P+I L L ++ S G I EIS L LDL
Sbjct: 1182 IGNFSSVQKLILDGNMFTGRIPPQIGRLQQLSKIDFSGNKFSGPIVPEISQCKLLTFLDL 1361
Query: 360 SMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASE 417
S N L G IP + + L ++ S N+L G +PS I S+ + +FSYNNL S
Sbjct: 1362 SRNELSGDIPNEITGMRILNYLNLSRNHLVGGIPSSI-SSMQSLTSVDFSYNNL----SG 1526
Query: 418 IKPDIMKTSFFGSVNSCPIAANPS----FFKKRRDVGHRGMKLALVLTLSLIFAL---AG 470
+ P + S+F N NP + +D G V LS F L G
Sbjct: 1527 LVPGTGQFSYF---NYTSFLGNPDLCGPYLGACKDGVANGAHQPHVKGLSSSFKLLLVVG 1697
Query: 471 ILF--LAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEK 528
+L +AF K +K+ S A + + F++
Sbjct: 1698 LLLCSIAFAVAAIFKARSLKKAS-------------------------GARAWKLTAFQR 1802
Query: 529 PLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGS--TLTDEEA 586
L+ T D+L ++ +G G VY+G +P HVAVK L S + D
Sbjct: 1803 --LDFTVDDVLHCLKE---DNIIGKGGAGIVYKGAMPNGDHVAVKRLPAMSRGSSHDHGF 1967
Query: 587 ARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTD 646
E++ LGRI+H ++V L G+C + + +Y+YM NG+L +L+
Sbjct: 1968 NAEIQTLGRIRHRHIVRLLGFCSNHETNLLVYEYMPNGSLGEVLH--------------- 2102
Query: 647 TWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDY 706
G +G W R+KIA+ A+ L +LHH CSP I+HR VK++++ LD
Sbjct: 2103 ------------GKKGGHLHWDTRYKIAVEAAKGLCYLHHDCSPLIVHRDVKSNNILLDS 2246
Query: 707 DLEPRLSDFGLAKIF-GSGLDE--EIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLF 763
+ E ++DFGLAK SG E GS GY+ PE++ KSDVY FGVVL
Sbjct: 2247 NHEAHVADFGLAKFLQDSGTSECMSAIAGSYGYIAPEYAYT--LKVDEKSDVYSFGVVLL 2420
Query: 764 ELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTS--RAIDPKICDTGSDEQIEEALKE 821
EL+TG+KPVG + + +V WVR + N+ + +DP++ E + +
Sbjct: 2421 ELITGRKPVG----EFGDGVDIVQWVRKMTDSNKEGVLKVLDPRLPSVPLHEVMH--VFY 2582
Query: 822 VGYLCTADLPFKRPTMQQIVGLLKDI 847
V LC + +RPTM+++V +L ++
Sbjct: 2583 VAMLCVEEQAVERPTMREVVQILTEL 2660
Score = 63.2 bits (152), Expect = 5e-10
Identities = 55/200 (27%), Positives = 93/200 (46%), Gaps = 5/200 (2%)
Frame = +3
Query: 228 SNFSRLKSIVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYL 285
++ + L + +L+++ N F G I + L L+LS N F +L L
Sbjct: 21 ADVAHLPFLSNLSLASNKFSGPIPPSLSALSGLRFLNLSNNVFNETFPSELSRLQNLEVL 200
Query: 286 DLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIP 345
DL N ++G + + NL+HL L N FS Q P+ L+YL +S L G IP
Sbjct: 201 DLYNNNMTGVLPLAVAQMQNLRHLHLGGNFFSGQIPPEYGRWQRLQYLAVSGNELEGTIP 380
Query: 346 DEISHLGNLNALDLS-MNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMK 402
EI +L +L L + N G IP + L +D ++ LSG +P+ +L K++
Sbjct: 381 PEIGNLSSLRELYIGYYNTYTGGIPPEIGNLSELVRLDAAYCGLSGEIPA----ALGKLQ 548
Query: 403 KYNFSYNNLTLCASEIKPDI 422
K + + + + + P++
Sbjct: 549 KLDTLFLQVNALSGSLTPEL 608
Score = 58.9 bits (141), Expect = 9e-09
Identities = 37/102 (36%), Positives = 55/102 (53%), Gaps = 2/102 (1%)
Frame = +3
Query: 292 LSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHL 351
LSG + ++ + L +LSLA N+FS P + L GL +LNLS P E+S L
Sbjct: 3 LSGPLSADVAHLPFLSNLSLASNKFSGPIPPSLSALSGLRFLNLSNNVFNETFPSELSRL 182
Query: 352 GNLNALDLSMNHLDGKIPL--LKNKHLQVIDFSHNNLSGPVP 391
NL LDL N++ G +PL + ++L+ + N SG +P
Sbjct: 183 QNLEVLDLYNNNMTGVLPLAVAQMQNLRHLHLGGNFFSGQIP 308
>TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, complete
Length = 3192
Score = 290 bits (742), Expect = 2e-78
Identities = 249/823 (30%), Positives = 379/823 (45%), Gaps = 39/823 (4%)
Frame = +3
Query: 64 NKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLS 122
N +V L+++ L+G IP +GKL KL +L L N ++ +L + +L SLKS++LS
Sbjct: 717 NLSELVRLDVAYCALSGEIP-AALGKLQKLDTLFLQVNALSGSLTPELGNLKSLKSMDLS 893
Query: 123 SNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGI 182
+N +SG + + G + +L +N IPE + L +L+V++L N SIP G+
Sbjct: 894 NNMLSGEIPASFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNLTGSIPEGL 1073
Query: 183 LKCQSLVSIDLSSNQLSGTLPHGF--GDAFPKLRTLN---------------------LA 219
K L +DLSSN+L+GTLP G+ L TL +
Sbjct: 1074 GKNGRLNLVDLSSNKLTGTLPPYLCSGNTLQTLITLGNFLFGPIPESLGTCESLTRIRMG 1253
Query: 220 ENNIYGGV-SNFSRLKSIVSLNISGNSFQGSIIEV--FVLKLEALDLSRNQFQGHISQVK 276
EN + G + L + + + N G EV + L + LS NQ G +S
Sbjct: 1254 ENFLNGSIPKGLFGLPKLTQVELQDNYLSGEFPEVGSVAVNLGQITLSNNQLSGALSPSI 1433
Query: 277 YNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLS 336
N+S + L L N +G I + L + + N+FS P+I L +L+LS
Sbjct: 1434 GNFSSVQKLLLDGNMFTGRIPTQIGRLQQLSKIDFSGNKFSGPIAPEISQCKLLTFLDLS 1613
Query: 337 KTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFI 394
+ L G IP+EI+ + LN L+LS NHL G IP + + L +DFS+NNLSG VP
Sbjct: 1614 RNELSGDIPNEITGMRILNYLNLSKNHLVGSIPSSISSMQSLTSVDFSYNNLSGLVPG-- 1787
Query: 395 LKSLPKMKKYNFSYNNLTLCASEIKPDIMKTSFFGSVNSCPIAANPSFFKKRRDVGHRGM 454
FSY N T PD+ A+ K +
Sbjct: 1788 --------TGQFSYFNYTSFLGN--PDLCGPYLGACKGGVANGAHQPHVKGLSS----SL 1925
Query: 455 KLALVLTL---SLIFALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTW 511
KL LV+ L S+ FA+A I K +K+ S
Sbjct: 1926 KLLLVVGLLLCSIAFAVAAIF----------KARSLKKAS-------------------- 2015
Query: 512 VADVKQATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVA 571
+A + + F++ L+ T D+L ++ +G G VY+G +P HVA
Sbjct: 2016 -----EARAWKLTAFQR--LDFTVDDVLHCLKE---DNIIGKGGAGIVYKGAMPNGDHVA 2165
Query: 572 VKVLVVGS--TLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNL 629
VK L S + D E++ LGRI+H ++V L G+C + + +Y+YM NG+L +
Sbjct: 2166 VKRLPAMSRGSSHDHGFNAEIQTLGRIRHRHIVRLLGFCSNHETNLLVYEYMPNGSLGEV 2345
Query: 630 LYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCS 689
L+ G +G W R+KIA+ A+ L +LHH CS
Sbjct: 2346 LH---------------------------GKKGGHLHWDTRYKIAVEAAKGLCYLHHDCS 2444
Query: 690 PPIIHRAVKASSVYLDYDLEPRLSDFGLAKIF-GSGLDE--EIARGSPGYVPPEFSQPEF 746
P I+HR VK++++ LD + E ++DFGLAK SG E GS GY+ PE++
Sbjct: 2445 PLIVHRDVKSNNILLDSNHEAHVADFGLAKFLQDSGTSECMSAIAGSYGYIAPEYAYT-- 2618
Query: 747 ESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTS--RAIDP 804
KSDVY FGVVL EL+TG+KPVG + + +V WVR + N+ + +DP
Sbjct: 2619 LKVDEKSDVYSFGVVLLELITGRKPVG----EFGDGVDIVQWVRKMTDSNKEGVLKVLDP 2786
Query: 805 KICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKDI 847
++ E + + V LC + +RPTM+++V +L ++
Sbjct: 2787 RLPSVPLHEVMH--VFYVAMLCVEEQAVERPTMREVVQILTEL 2909
Score = 137 bits (346), Expect = 2e-32
Identities = 128/421 (30%), Positives = 185/421 (43%), Gaps = 39/421 (9%)
Frame = +3
Query: 10 LVLTFLFKHLASQQPNTDEFYVSEFLKKMGLTSSS--------KVYNFSSSVCSWKGVYC 61
++ FLF H P T +SE+ + L S +N S CSW GV C
Sbjct: 33 VLFVFLFFHF--HFPETLSAPISEYRALLSLRSVITDATPPVLSSWNASIPYCSWLGVTC 206
Query: 62 DSNKEHVVELNLSGIGLTG------------------------PIPDTTIGKLNKLHSLD 97
D N+ HV LNL+G+ L+G PIP + + L+ L L+
Sbjct: 207 D-NRRHVTALNLTGLDLSGTLSADVAHLPFLSNLSLAANKFSGPIPPS-LSALSGLRYLN 380
Query: 98 LSNNKIT-TLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPE 156
LSNN T PS+ W L SL+ L+L +N+++G L P
Sbjct: 381 LSNNVFNETFPSELWRLQSLEVLDLYNNNMTGVL------------------------PL 488
Query: 157 ALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTL 216
A++ + +L+ L L N F IP + Q L + +S N+L GT+P G+ LR L
Sbjct: 489 AVAQMQNLRHLHLGGNFFSGQIPPEYGRWQRLQYLAVSGNELDGTIPPEIGN-LTSLREL 665
Query: 217 NLAENNIY-GGV-SNFSRLKSIVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHI 272
+ N Y GG+ L +V L+++ + G I + KL+ L L N G +
Sbjct: 666 YIGYYNTYTGGIPPEIGNLSELVRLDVAYCALSGEIPAALGKLQKLDTLFLQVNALSGSL 845
Query: 273 SQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEY 332
+ N L +DLS N LSGEI + N+ L+L N+ I L LE
Sbjct: 846 TPELGNLKSLKSMDLSNNMLSGEIPASFGELKNITLLNLFRNKLHGAIPEFIGELPALEV 1025
Query: 333 LNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPV 390
+ L + +L G IP+ + G LN +DLS N L G +P L LQ + N L GP+
Sbjct: 1026VQLWENNLTGSIPEGLGKNGRLNLVDLSSNKLTGTLPPYLCSGNTLQTLITLGNFLFGPI 1205
Query: 391 P 391
P
Sbjct: 1206P 1208
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 287 bits (734), Expect = 2e-77
Identities = 249/816 (30%), Positives = 392/816 (47%), Gaps = 32/816 (3%)
Frame = +1
Query: 64 NKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLS 122
N ++V L+ + GL+G IP +GKL L +L L N ++ +L + SL SLKS++LS
Sbjct: 784 NLSNLVRLDAAYCGLSGEIP-AELGKLQNLDTLFLQVNALSGSLTPELGSLKSLKSMDLS 960
Query: 123 SNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGI 182
+N +SG + + L +L +N IPE + L +L+VL+L N F SIP +
Sbjct: 961 NNMLSGEVPASFAELKNLTLLNLFRNKLHGAIPEFVGELPALEVLQLWENNFTGSIPQNL 1140
Query: 183 LKCQSLVSIDLSSNQLSGTLPHG--FGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSL 239
L +DLSSN+++GTLP +G+ +L+TL N ++G + + + KS+ +
Sbjct: 1141 GNNGRLTLVDLSSNKITGTLPPNMCYGN---RLQTLITLGNYLFGPIPDSLGKCKSLNRI 1311
Query: 240 NISGNSFQGSIIE-VFVL-KLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIF 297
+ N GSI + +F L KL ++L N G + + L + LS NQLSG +
Sbjct: 1312 RMGENFLNGSIPKGLFGLPKLTQVELQDNLLTGQFPEDGSIATDLGQISLSNNQLSGSLP 1491
Query: 298 QNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNAL 357
+ N +++ L L N F+ + P+I ML L ++ S G I EIS L +
Sbjct: 1492 STIGNFTSMQKLLLNGNEFTGRIPPQIGMLQQLSKIDFSHNKFSGPIAPEISKCKLLTFI 1671
Query: 358 DLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCA 415
DLS N L G+IP + + L ++ S N+L G +P I S+ + +FSYNN
Sbjct: 1672 DLSGNELSGEIPNKITSMRILNYLNLSRNHLDGSIPGNIA-SMQSLTSVDFSYNNF---- 1836
Query: 416 SEIKPDIMK------TSFFGSVNSCPIAANP---SFFKKRRDVGHRG-----MKLALVLT 461
S + P + TSF G+ C P R +G +KL LV+
Sbjct: 1837 SGLVPGTGQFGYFNYTSFLGNPELCGPYLGPCKDGVANGPRQPHVKGPFSSSLKLLLVIG 2016
Query: 462 L---SLIFALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQA 518
L S++FA+A I K +K+ S +A
Sbjct: 2017 LLVCSILFAVAAIF----------KARALKKAS-------------------------EA 2091
Query: 519 TSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVG 578
+ + F++ L+ T D+L ++ +G G VY+G +P +VAVK L
Sbjct: 2092 RAWKLTAFQR--LDFTVDDVLDCLKE---DNIIGKGGAGIVYKGAMPNGDNVAVKRLPAM 2256
Query: 579 S--TLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLG 636
S + D E++ LGRI+H ++V L G+C + + +Y+YM NG+L +L+
Sbjct: 2257 SRGSSHDHGFNAEIQTLGRIRHRHIVRLLGFCSNHETNLLVYEYMPNGSLGEVLH----- 2421
Query: 637 VQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRA 696
G +G W R+KIA+ A+ L +LHH CSP I+HR
Sbjct: 2422 ----------------------GKKGGHLHWDTRYKIAVEAAKGLCYLHHDCSPLIVHRD 2535
Query: 697 VKASSVYLDYDLEPRLSDFGLAKIF-GSGLDE--EIARGSPGYVPPEFSQPEFESPTPKS 753
VK++++ LD + E ++DFGLAK SG E GS GY+ PE++ KS
Sbjct: 2536 VKSNNILLDSNFEAHVADFGLAKFLQDSGASECMSAIAGSYGYIAPEYAYT--LKVDEKS 2709
Query: 754 DVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTS--RAIDPKICDTGS 811
DVY FGVVL EL+TG+KPVG + + +V WVR + N+ + +D ++
Sbjct: 2710 DVYSFGVVLLELVTGRKPVG----EFGDGVDIVQWVRKMTDSNKEGVLKVLDSRLPSVPL 2877
Query: 812 DEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKDI 847
E + + V LC + +RPTM+++V +L ++
Sbjct: 2878 HEVMH--VFYVAMLCVEEQAVERPTMREVVQILTEL 2979
Score = 152 bits (384), Expect = 6e-37
Identities = 128/408 (31%), Positives = 194/408 (47%), Gaps = 11/408 (2%)
Frame = +1
Query: 9 VLVLTFLFKHLASQQPNTDEFYVSEFLKKMGLTSSS----KVYNFSSSVCSWKGVYCDSN 64
VLVL FLF H + Q E+ K LT +N S+ CSW G+ CDS
Sbjct: 106 VLVLFFLFLH-SLQAARISEYRALLSFKASSLTDDPTHALSSWNSSTPFCSWFGLTCDSR 282
Query: 65 KEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSS 123
+ HV LNL+ + L+G + D + L L L L++NK + +P+ F +L++L+ LNLS+
Sbjct: 283 R-HVTSLNLTSLSLSGTLSDD-LSHLPFLSHLSLADNKFSGPIPASFSALSALRFLNLSN 456
Query: 124 NHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGIL 183
N + + + + LE DL N+ + E+P +++++ L+ L L N F IP
Sbjct: 457 NVFNATFPSQLNRLANLEVLDLYNNNMTGELPLSVAAMPLLRHLHLGGNFFSGQIPPEYG 636
Query: 184 KCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGG--VSNFSRLKSIVSLNI 241
Q L + LS N+L+GT+ G+ LR L + N Y G L ++V L+
Sbjct: 637 TWQHLQYLALSGNELAGTIAPELGN-LSSLRELYIGYYNTYSGGIPPEIGNLSNLVRLDA 813
Query: 242 SGNSFQGSI-IEVFVLK-LEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQN 299
+ G I E+ L+ L+ L L N G ++ + L +DLS N LSGE+ +
Sbjct: 814 AYCGLSGEIPAELGKLQNLDTLFLQVNALSGSLTPELGSLKSLKSMDLSNNMLSGEVPAS 993
Query: 300 LNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDL 359
NL L+L N+ + L LE L L + + G IP + + G L +DL
Sbjct: 994 FAELKNLTLLNLFRNKLHGAIPEFVGELPALEVLQLWENNFTGSIPQNLGNNGRLTLVDL 1173
Query: 360 SMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYN 405
S N + G +P + LQ + N L GP+P SL K K N
Sbjct: 1174SSNKITGTLPPNMCYGNRLQTLITLGNYLFGPIPD----SLGKCKSLN 1305
>TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment), complete
Length = 3298
Score = 263 bits (671), Expect = 3e-70
Identities = 233/804 (28%), Positives = 378/804 (46%), Gaps = 28/804 (3%)
Frame = +1
Query: 71 LNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISGS 129
L+LS L+G IP + + L L +L L N +T T+PS+ ++ SL SL+LS N ++G
Sbjct: 832 LDLSSCNLSGEIPPS-LANLTNLDTLFLQINNLTGTIPSELSAMVSLMSLDLSINDLTGE 1008
Query: 130 LTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLV 189
+ + L + +N+ +P + L +L+ L+L N F +P + + L
Sbjct: 1009 IPMSFSQLRNLTLMNFFQNNLRGSVPSFVGELPNLETLQLWDNNFSFVLPPNLGQNGKLK 1188
Query: 190 SIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSN-FSRLKSIVSLNISGNSFQG 248
D+ N +G +P + +L+T+ + +N G + N KS+ + S N G
Sbjct: 1189 FFDVIKNHFTGLIPRDLCKS-GRLQTIMITDNFFRGPIPNEIGNCKSLTKIRASNNYLNG 1365
Query: 249 SIIEVFVLKLEA---LDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMN 305
++ + KL + ++L+ N+F G + + + L L LS N SG+I L N
Sbjct: 1366 -VVPSGIFKLPSVTIIELANNRFNGELPP-EISGESLGILTLSNNLFSGKIPPALKNLRA 1539
Query: 306 LKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLD 365
L+ LSL N F + ++ L L +N+S +L G IP ++ +L A+DLS N L+
Sbjct: 1540 LQTLSLDANEFVGEIPGEVFDLPMLTVVNISGNNLTGPIPTTLTRCVSLTAVDLSRNMLE 1719
Query: 366 GKIPL-LKN-KHLQVIDFSHNNLSGPVPSFI--LKSLPKMKKYNFSYNNLTLCASEIKPD 421
GKIP +KN L + + S N +SGPVP I + SL + N ++ +
Sbjct: 1720 GKIPKGIKNLTDLSIFNVSINQISGPVPEEIRFMLSLTTLDLSNNNFIGKVPTGGQFAV- 1896
Query: 422 IMKTSFFGSVNSCPIAANPSF-------FKKRRDVGHRGMKLALVLTLSLIFALAGILF- 473
+ SF G+ N C + P+ KKRR G +K V+ + + A +L
Sbjct: 1897 FSEKSFAGNPNLCTSHSCPNSSLYPDDALKKRR--GPWSLKSTRVIVIVIALGTAALLVA 2070
Query: 474 LAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKPLLNI 533
+ R+ KM K TW + F++ LN
Sbjct: 2071 VTVYMMRRRKMNLAK---------------------TW----------KLTAFQR--LNF 2151
Query: 534 TFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLV-VGSTLTDEEAARELEF 592
D++ + ++ +G G VYRG +P VA+K LV GS D E+E
Sbjct: 2152 KAEDVVECLKEEN---IIGKGGAGIVYRGSMPNGTDVAIKRLVGAGSGRNDYGFKAEIET 2322
Query: 593 LGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEAD 652
LG+I+H N++ L GY + + +Y+YM NG+L L+
Sbjct: 2323 LGKIRHRNIMRLLGYVSNKETNLLLYEYMPNGSLGEWLH--------------------- 2439
Query: 653 NGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRL 712
G++G W R+KIA+ A+ L +LHH CSP IIHR VK++++ LD DLE +
Sbjct: 2440 ------GAKGGHLKWEMRYKIAVEAAKGLCYLHHDCSPLIIHRDVKSNNILLDGDLEAHV 2601
Query: 713 SDFGLAK-IFGSGLDEEIA--RGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGK 769
+DFGLAK ++ G + ++ GS GY+ PE++ KSDVY FGVVL EL+ G+
Sbjct: 2602 ADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYT--LKVDEKSDVYSFGVVLLELIIGR 2775
Query: 770 KPVGDDYTDDKEATTLVSWV-RGLVRKNQTSRA------IDPKICDTGSDEQIEEALKEV 822
KPVG + + +V WV + + Q S A +DP++ +G + +
Sbjct: 2776 KPVG----EFGDGVDIVGWVNKTRLELAQPSDAALVLAVVDPRL--SGYPLTSVIYMFNI 2937
Query: 823 GYLCTADLPFKRPTMQQIVGLLKD 846
+C ++ RPTM+++V +L +
Sbjct: 2938 AMMCVKEMGPARPTMREVVHMLSE 3009
Score = 119 bits (298), Expect = 6e-27
Identities = 105/372 (28%), Positives = 172/372 (46%), Gaps = 4/372 (1%)
Frame = +1
Query: 45 KVYNFSSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT 104
K + S+ C + GV CD + VV +N+S + L G +P IG+L+KL
Sbjct: 247 KFFPSLSAHCFFSGVKCD-RELRVVAINVSFVPLFGHLPPE-IGQLDKL----------- 387
Query: 105 TLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIP-EALSSLVS 163
++L +S N+++G L + L++ ++S N FS P + + +
Sbjct: 388 ------------ENLTVSQNNLTGVLPKELAALTSLKHLNISHNVFSGHFPGQIILPMTK 531
Query: 164 LKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNI 223
L+VL + N F +P ++K + L + L N SG++P + + F L L+L+ N++
Sbjct: 532 LEVLDVYDNNFTGPLPVELVKLEKLKYLKLDGNYFSGSIPESYSE-FKSLEFLSLSTNSL 708
Query: 224 YGGV-SNFSRLKSIVSLNISGNSFQGSIIEVFVLKLEALDLSRNQFQGHISQVKYNWSHL 282
G + + S+LK++ L + N N ++G I + L
Sbjct: 709 SGKIPKSLSKLKTLRYLKLGYN---------------------NAYEGGIPPEFGSMKSL 825
Query: 283 VYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVG 342
YLDLS LSGEI +L N NL L L N +L G
Sbjct: 826 RYLDLSSCNLSGEIPPSLANLTNLDTLFLQIN------------------------NLTG 933
Query: 343 HIPDEISHLGNLNALDLSMNHLDGKIPL--LKNKHLQVIDFSHNNLSGPVPSFILKSLPK 400
IP E+S + +L +LDLS+N L G+IP+ + ++L +++F NNL G VPSF+ LP
Sbjct: 934 TIPSELSAMVSLMSLDLSINDLTGEIPMSFSQLRNLTLMNFFQNNLRGSVPSFV-GELPN 1110
Query: 401 MKKYNFSYNNLT 412
++ NN +
Sbjct: 1111LETLQLWDNNFS 1146
Score = 62.0 bits (149), Expect = 1e-09
Identities = 58/191 (30%), Positives = 94/191 (48%), Gaps = 9/191 (4%)
Frame = +1
Query: 256 LKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNR 315
L++ A+++S GH+ L L +S+N L+G + + L +LKHL+++ N
Sbjct: 307 LRVVAINVSFVPLFGHLPPEIGQLDKLENLTVSQNNLTGVLPKELAALTSLKHLNISHNV 486
Query: 316 FSRQKFPK--IEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP--LL 371
FS FP I + LE L++ + G +P E+ L L L L N+ G IP
Sbjct: 487 FSGH-FPGQIILPMTKLEVLDVYDNNFTGPLPVELVKLEKLKYLKLDGNYFSGSIPESYS 663
Query: 372 KNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNF---SYNNLTLCASEIKPDI--MKTS 426
+ K L+ + S N+LSG +P KSL K+K + YNN I P+ MK+
Sbjct: 664 EFKSLEFLSLSTNSLSGKIP----KSLSKLKTLRYLKLGYNN--AYEGGIPPEFGSMKSL 825
Query: 427 FFGSVNSCPIA 437
+ ++SC ++
Sbjct: 826 RYLDLSSCNLS 858
>TC204550 UP|Q8L3Y5 (Q8L3Y5) Receptor-like kinase RHG1, complete
Length = 2659
Score = 262 bits (669), Expect = 5e-70
Identities = 240/853 (28%), Positives = 384/853 (44%), Gaps = 28/853 (3%)
Frame = +2
Query: 28 EFYVSEFLKKMGLTSSSKVYNFSSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTI 87
E + E G S + + W G+ C + V+ + L GL G I D I
Sbjct: 329 EAFKQELADPEGFLRSWNDSGYGACSGGWVGIKCAQGQ--VIVIQLPWKGLRGRITDK-I 499
Query: 88 GKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLS 146
G+L L L L +N+I ++PS L +L+ + L +N ++GS+ ++G LL++ DLS
Sbjct: 500 GQLQGLRKLSLHDNQIGGSIPSTLGLLPNLRGVQLFNNRLTGSIPLSLGFCPLLQSLDLS 679
Query: 147 KNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGF 206
N + IP +L++ L L L N F +P+ + SL + L +N LSG+LP+ +
Sbjct: 680 NNLLTGAIPYSLANSTKLYWLNLSFNSFSGPLPASLTHSFSLTFLSLQNNNLSGSLPNSW 859
Query: 207 G----DAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISGNSFQGSII-EVFVL-KLE 259
G + F +L+ L L N G V ++ L+ + +++S N F G+I E+ L +L+
Sbjct: 860 GGNSKNGFFRLQNLILDHNFFTGDVPASLGSLRELNEISLSHNKFSGAIPNEIGTLSRLK 1039
Query: 260 ALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQ 319
LD+S N G++ N S L L+ N L +I Q+L NL L L+ N+FS
Sbjct: 1040 TLDISNNALNGNLPATLSNLSSLTLLNAENNLLDNQIPQSLGRLRNLSVLILSRNQFS-- 1213
Query: 320 KFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIPLL--KNKHLQ 377
GHIP I+++ +L LDLS+N+ G+IP+ + L
Sbjct: 1214 ----------------------GHIPSSIANISSLRQLDLSLNNFSGEIPVSFDSQRSLN 1327
Query: 378 VIDFSHNNLSGPVPSFILKSLPKMKKYNFS--YNNLTLCASEIKPDIMKTSFFGSVNSCP 435
+ + S+N+LSG VP + KK+N S N+ LC T F S
Sbjct: 1328 LFNVSYNSLSGSVPPLL------AKKFNSSSFVGNIQLCGYS-----PSTPCFSQAPSQG 1474
Query: 436 IAANPSFFKKRRDVGHRGMKLALVLTLSLIFALAGILF---------LAFGCRRKNKMWE 486
+ A P K H KL+ T +I +AG+L L F RK +
Sbjct: 1475 VIAPPPEVSKH----HHHRKLS---TKDIILVVAGVLLVVLIILCCVLLFCLIRKRSTSK 1633
Query: 487 VKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVP--VVIFEKPLLNITFADLLSATSN 544
G E + ++ ++ S+ W +KQ + +V F+ P+ T DLL AT+
Sbjct: 1634 AGNGQATEGRAVTYEDRKRSPSSCWWLMLKQVGRLEGNLVHFDGPMA-FTADDLLCATAE 1810
Query: 545 FDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFLGRIKHPNLVPL 604
++ + G VY+ L VAVK L E E+ LG+I+HPN++ L
Sbjct: 1811 -----IMGKSTNGTVYKAILEDGSQVAVKRLREKIAKGHREFESEVSVLGKIRHPNVLAL 1975
Query: 605 TGYCVA-GDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGL 663
Y + +++ ++DYM G+L + L+ G ++ DW T
Sbjct: 1976 RAYYLGPKGEKLLVFDYMSKGSLASFLHG--GGTETFIDWPT------------------ 2095
Query: 664 LTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGS 723
R KIA AR L LH IIH + +S+V LD + +++DFGL+++ +
Sbjct: 2096 ------RMKIAQDLARGLFCLH--SQENIIHGNLTSSNVLLDENTNAKIADFGLSRLMST 2251
Query: 724 GLDEEI--ARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKE 781
+ + G+ GY PE S + + K+D+Y GV+L ELLT K P
Sbjct: 2252 TANSNVIATAGALGYRAPELS--KLKKANTKTDIYSLGVILLELLTRKSP-----GVSMN 2410
Query: 782 ATTLVSWVRGLVRKNQTSRAIDPKICDTGS--DEQIEEALKEVGYLCTADLPFKRPTMQQ 839
L WV +V++ T+ D + S +++ LK + C P RP + Q
Sbjct: 2411 GLDLPQWVASVVKEEWTNEVFDADLMRDASTVGDELLNTLK-LALHCVDPSPSARPEVHQ 2587
Query: 840 IVGLLKDIEPTTS 852
++ L++I P S
Sbjct: 2588 VLQQLEEIRPERS 2626
>NP205681 receptor protein kinase-like protein
Length = 2946
Score = 258 bits (658), Expect = 1e-68
Identities = 227/798 (28%), Positives = 355/798 (44%), Gaps = 14/798 (1%)
Frame = +1
Query: 63 SNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNL 121
S+ ++ L+LS GLTG IP T +L L ++ +N + ++PS L +L++L L
Sbjct: 862 SDMVSLMSLDLSFNGLTGEIP-TRFSQLKNLTLMNFFHNNLRGSVPSFVGELPNLETLQL 1038
Query: 122 SSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSG 181
N+ S L N+G G + FD++KN FS IP L L+ + N F IP+
Sbjct: 1039 WENNFSSELPQNLGQNGKFKFFDVTKNHFSGLIPRDLCKSGRLQTFLITDNFFHGPIPNE 1218
Query: 182 ILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSNFSRLKSIVSLNI 241
I C+SL I S+N L+G +P G P + + LA N G + S+ L +
Sbjct: 1219 IANCKSLTKIRASNNYLNGAVPSGI-FKLPSVTIIELANNRFNGELPPEISGDSLGILTL 1395
Query: 242 SGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQN 299
S N F G I + L+ L L N+F G I ++ L +++S N L+G I
Sbjct: 1396 SNNLFTGKIPPALKNLRALQTLSLDTNEFLGEIPGEVFDLPMLTVVNISGNNLTGPIPTT 1575
Query: 300 LNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDL 359
++L + L+ N + ++ L L N+S + G +PDEI + +L LDL
Sbjct: 1576 FTRCVSLAAVDLSRNMLDGEIPKGMKNLTDLSIFNVSINQISGSVPDEIRFMLSLTTLDL 1755
Query: 360 SMNHLDGKIPLLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASEIK 419
S N+ GK+P Q + FS + +G N LC+S
Sbjct: 1756 SYNNFIGKVP----TGGQFLVFSDKSFAG---------------------NPNLCSS--- 1851
Query: 420 PDIMKTSFFGSVNSCPIAANPSFFKKRRDVGHRGMKLALVLTLSLIFALAGILFLAFGCR 479
+SCP S KKRR G +K V+ + + A A IL
Sbjct: 1852 ------------HSCP----NSSLKKRR--GPWSLKSTRVIVMVIALATAAILVAGTEYM 1977
Query: 480 RKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKPLLNITFADLL 539
R+ + +K A + + F++ LN+ +++
Sbjct: 1978 RRRR------------------------------KLKLAMTWKLTGFQR--LNLKAEEVV 2061
Query: 540 SATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLV-VGSTLTDEEAARELEFLGRIKH 598
++ +G G VYRG + VA+K LV GS D E+E +G+I+H
Sbjct: 2062 EC---LKEENIIGKGGAGIVYRGSMRNGSDVAIKRLVGAGSGRNDYGFKAEIETVGKIRH 2232
Query: 599 PNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNV 658
N++ L GY + + +Y+YM NG+L L+
Sbjct: 2233 RNIMRLLGYVSNKETNLLLYEYMPNGSLGEWLH--------------------------- 2331
Query: 659 GSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLA 718
G++G W R+KIA+ A+ L +LHH CSP IIHR VK++++ LD E ++DFGLA
Sbjct: 2332 GAKGGHLKWEMRYKIAVEAAKGLCYLHHDCSPLIIHRDVKSNNILLDAHFEAHVADFGLA 2511
Query: 719 KI---FGSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDD 775
K GS GS GY+ PE++ KSDVY FGVVL EL+ G+KPVG
Sbjct: 2512 KFLYDLGSSQSMSSIAGSYGYIAPEYAYT--LKVDEKSDVYSFGVVLLELIIGRKPVG-- 2679
Query: 776 YTDDKEATTLVSWV-RGLVRKNQTSRA------IDPKICDTGSDEQIEEALKEVGYLCTA 828
+ + +V WV + + +Q S A +DP++ +G + + +C
Sbjct: 2680 --EFGDGVDIVGWVNKTRLELSQPSDAAVVLAVVDPRL--SGYPLISVIYMFNIAMMCVK 2847
Query: 829 DLPFKRPTMQQIVGLLKD 846
++ RPTM+++V +L +
Sbjct: 2848 EVGPTRPTMREVVHMLSN 2901
Score = 155 bits (393), Expect = 5e-38
Identities = 127/372 (34%), Positives = 180/372 (48%), Gaps = 13/372 (3%)
Frame = +1
Query: 47 YNFSSSV---CSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKI 103
+ FS+S+ C + GV CD VV +N+S + L G +P IG+L+KL +L +S N +
Sbjct: 154 WKFSTSLSAHCFFSGVSCDQELR-VVAINVSFVPLFGHVPPE-IGELDKLENLTISQNNL 327
Query: 104 T-TLPSDFWSLTSLKSLNLSSNHISGSLTNNIG-NFGLLENFDLSKNSFSDEIPEALSSL 161
T LP + +LTSLK LN+S N SG I LE D+ N+F+ +PE L
Sbjct: 328 TGELPKELAALTSLKHLNISHNVFSGYFPGKIILPMTELEVLDVYDNNFTGSLPEEFVKL 507
Query: 162 VSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAEN 221
LK LKLD N F SIP + +SL + LS+N LSG +P LR L L N
Sbjct: 508 EKLKYLKLDGNYFSGSIPESYSEFKSLEFLSLSTNSLSGNIPKSL-SKLKTLRILKLGYN 684
Query: 222 NIY-GGV-SNFSRLKSIVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVKY 277
N Y GG+ F ++S+ L++S + G I + L+ L L N G I
Sbjct: 685 NAYEGGIPPEFGTMESLKYLDLSSCNLSGEIPPSLANMRNLDTLFLQMNNLTGTIPSELS 864
Query: 278 NWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSK 337
+ L+ LDLS N L+GEI + NL ++ N + L LE L L +
Sbjct: 865 DMVSLMSLDLSFNGLTGEIPTRFSQLKNLTLMNFFHNNLRGSVPSFVGELPNLETLQLWE 1044
Query: 338 TSLVGHIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFI- 394
+ +P + G D++ NH G IP L K+ LQ + N GP+P+ I
Sbjct: 1045NNFSSELPQNLGQNGKFKFFDVTKNHFSGLIPRDLCKSGRLQTFLITDNFFHGPIPNEIA 1224
Query: 395 -LKSLPKMKKYN 405
KSL K++ N
Sbjct: 1225NCKSLTKIRASN 1260
>TC216858 homologue to UP|Q76FZ8 (Q76FZ8) Brassinosteroid receptor, partial
(29%)
Length = 1320
Score = 230 bits (586), Expect = 2e-60
Identities = 138/338 (40%), Positives = 196/338 (57%), Gaps = 7/338 (2%)
Frame = +1
Query: 518 ATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVV 577
A S+ + FEKPL +TFADLL AT+ F +L+ G FG VY+ L VA+K L+
Sbjct: 1 ALSIFLATFEKPLRKLTFADLLDATNGFHNDSLIGSGGFGDVYKAQLKDGSVVAIKKLIH 180
Query: 578 GSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGV 637
S D E E+E +G+IKH NLVPL GYC G++R+ +Y+YM+ G+L+++L+D
Sbjct: 181 VSGQGDREFTAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDQK--- 351
Query: 638 QSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAV 697
G+ W R KIA+G AR LAFLHH C P IIHR +
Sbjct: 352 ----------------------KAGIKLNWAIRRKIAIGAARGLAFLHHNCIPHIIHRDM 465
Query: 698 KASSVYLDYDLEPRLSDFGLAKIFGSGLDEEIA----RGSPGYVPPEFSQPEFESPTPKS 753
K+S+V LD +LE R+SDFG+A++ S +D ++ G+PGYVPPE+ Q F T K
Sbjct: 466 KSSNVLLDENLEARVSDFGMARLM-SAMDTHLSVSTLAGTPGYVPPEYYQ-SFRCST-KG 636
Query: 754 DVYCFGVVLFELLTGKKPVGD-DYTDDKEATTLVSWVRGLVRKNQTSRAIDPKIC--DTG 810
DVY +GVVL ELLTGK+P D+ D+ LV WV+ K + S DP++ D
Sbjct: 637 DVYSYGVVLLELLTGKRPTDSADFGDN----NLVGWVKQHA-KLKISDIFDPELMKEDPN 801
Query: 811 SDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKDIE 848
+ ++ + LK + C D P++RPTM Q++ + K+I+
Sbjct: 802 LEMELLQHLK-IAVSCLDDRPWRRPTMIQVMAMFKEIQ 912
>TC209224 similar to UP|BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor
(Brassinosteroid LRR receptor kinase) , partial (14%)
Length = 880
Score = 188 bits (478), Expect = 8e-48
Identities = 116/274 (42%), Positives = 161/274 (58%), Gaps = 13/274 (4%)
Frame = +3
Query: 589 ELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTW 648
E+E LG+IKH NLVPL GYC G++R+ +Y+YME G+L+ +L+ G T D TW
Sbjct: 6 EMETLGKIKHRNLVPLLGYCKVGEERLLVYEYMEYGSLEEMLH----GRIKTRDRRILTW 173
Query: 649 EEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDL 708
EE R KIA G A+ L FLHH C P IIHR +K+S+V LD+++
Sbjct: 174 EE-------------------RKKIARGAAKGLCFLHHNCIPHIIHRDMKSSNVLLDHEM 296
Query: 709 EPRLSDFGLAKIFGSGLDEEIA----RGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFE 764
E R+SDFG+A++ S LD ++ G+PGYVPPE+ Q F T K DVY FGVV+ E
Sbjct: 297 ESRVSDFGMARLI-SALDTHLSVSTLAGTPGYVPPEYYQ-SFRC-TVKGDVYSFGVVMLE 467
Query: 765 LLTGKKPVG-DDYTDDKEATTLVSWVRGLVRKNQTSRAIDPK--ICDTGSDEQIEEALKE 821
LL+GK+P +D+ D T LV W + VR+ + ID + G+DE + +KE
Sbjct: 468 LLSGKRPTDKEDFGD----TNLVGWAKIKVREGKQMEVIDNDLLLATQGTDEAEAKEVKE 635
Query: 822 -VGYL-----CTADLPFKRPTMQQIVGLLKDIEP 849
+ YL C DLP +RP M Q+V +L+++ P
Sbjct: 636 MIRYLEITLQCVDDLPSRRPNMLQVVAMLRELMP 737
>TC229775 weakly similar to UP|KPEL_DROME (Q05652) Probable
serine/threonine-protein kinase pelle , partial (8%)
Length = 1250
Score = 169 bits (427), Expect = 6e-42
Identities = 109/316 (34%), Positives = 158/316 (49%), Gaps = 3/316 (0%)
Frame = +1
Query: 534 TFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFL 593
TF +L+ T+NF R +L G FG VY GF+ + VAVK+L + E+ E++ L
Sbjct: 52 TFNELVKITNNFTR--ILGRGGFGKVYHGFID-DTQVAVKMLSPSAVRGYEQFLAEVKLL 222
Query: 594 GRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADN 653
R+ H NL L GYC + IY+YM NGNL +
Sbjct: 223 MRVHHRNLTSLVGYCNEENNIGLIYEYMANGNLDEI------------------------ 330
Query: 654 GIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLS 713
+ S TW R +IAL A+ L +LH+GC PPIIHR VK +++ L+ + + +L+
Sbjct: 331 -VSGKSSRAKFLTWEDRLQIALDAAQGLEYLHNGCKPPIIHRDVKCANILLNENFQAKLA 507
Query: 714 DFGLAKIF---GSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKK 770
DFGL+K F G + G+PGY+ PE+S T KSDVY FGVVL E++TG+
Sbjct: 508 DFGLSKSFPTDGGSYMSTVVAGTPGYLDPEYSIS--SRLTEKSDVYSFGVVLLEMVTGQP 681
Query: 771 PVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTADL 830
+ T DK T + WV+ ++ D + + + + E+G +
Sbjct: 682 AIAK--TPDK--THISQWVKSMLSNGDIKNIADSRFKEDFDTSSVWR-IVEIGMASVSIS 846
Query: 831 PFKRPTMQQIVGLLKD 846
PFKRP+M IV LK+
Sbjct: 847 PFKRPSMSYIVNELKE 894
>TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, partial (38%)
Length = 1009
Score = 166 bits (420), Expect = 4e-41
Identities = 112/315 (35%), Positives = 161/315 (50%), Gaps = 3/315 (0%)
Frame = +3
Query: 534 TFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFL 593
TF++L AT F + LAEG FG V+RG LP +AVK + ST D+E E+E L
Sbjct: 9 TFSELQLATGGFSQANFLAEGGFGSVHRGVLPDGQVIAVKQYKLASTQGDKEFCSEVEVL 188
Query: 594 GRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADN 653
+H N+V L G+CV +R+ +Y+Y+ NG+L + +Y
Sbjct: 189 SCAQHRNVVMLIGFCVDDGRRLLVYEYICNGSLDSHIY---------------------R 305
Query: 654 GIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPP-IIHRAVKASSVYLDYDLEPRL 712
QNV W R KIA+G AR L +LH C I+HR ++ +++ L +D E +
Sbjct: 306 RKQNV------LEWSARQKIAVGAARGLRYLHEECRVGCIVHRDMRPNNILLTHDFEALV 467
Query: 713 SDFGLAKIFGSG--LDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKK 770
DFGLA+ G E G+ GY+ PE++Q T K+DVY FG+VL EL+TG+K
Sbjct: 468 GDFGLARWQPDGDMGVETRVIGTFGYLAPEYAQS--GQITEKADVYSFGIVLLELVTGRK 641
Query: 771 PVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTADL 830
V D K L W R L+ K T + IDP + + D+++ LK LC
Sbjct: 642 AV--DINRPKGQQCLSEWARPLLEKQATYKLIDPSLRNCYVDQEVYRMLK-CSSLCIGRD 812
Query: 831 PFKRPTMQQIVGLLK 845
P RP M Q++ +L+
Sbjct: 813 PHLRPRMSQVLRMLE 857
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 165 bits (418), Expect = 7e-41
Identities = 125/394 (31%), Positives = 194/394 (48%), Gaps = 5/394 (1%)
Frame = +3
Query: 456 LALVLTLSLIFALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADV 515
+ + +T++++ + GI FL+ R+K +QGS +E
Sbjct: 651 IVVPITVAVLIFIVGICFLSRRARKK------QQGSVKEG-------------------- 752
Query: 516 KQATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVL 575
K A +P V L F+ + +AT+ F L EG FG VY+G L VAVK L
Sbjct: 753 KTAYDIPTV----DSLQFDFSTIEAATNKFSADNKLGEGGFGEVYKGTLSSGQVVAVKRL 920
Query: 576 VVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPL 635
S EE E+ + +++H NLV L G+C+ G+++I +Y+Y+ N +L +L+D
Sbjct: 921 SKSSGQGGEEFKNEVVVVAKLQHRNLVRLLGFCLQGEEKILVYEYVPNKSLDYILFD--- 1091
Query: 636 GVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHR 695
+ E D W R+KI G AR + +LH IIHR
Sbjct: 1092---------PEKQRELD--------------WGRRYKIIGGIARGIQYLHEDSRLRIIHR 1202
Query: 696 AVKASSVYLDYDLEPRLSDFGLAKIFG----SGLDEEIARGSPGYVPPEFS-QPEFESPT 750
+KAS++ LD D+ P++SDFG+A+IFG G I G+ GY+ PE++ EF +
Sbjct: 1203DLKASNILLDGDMNPKISDFGMARIFGVDQTQGNTSRIV-GTYGYMAPEYAMHGEF---S 1370
Query: 751 PKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTG 810
KSDVY FGV+L E+L+GKK TD E L+S+ L + +DP + ++
Sbjct: 1371VKSDVYSFGVLLMEILSGKKNSSFYQTDGAE--DLLSYAWQLWKDGTPLELMDPILRESY 1544
Query: 811 SDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLL 844
+ ++ ++ +G LC + P RPTM IV +L
Sbjct: 1545NQNEVIRSI-HIGLLCVQEDPADRPTMATIVLML 1643
>TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicited protein
264, partial (73%)
Length = 1047
Score = 165 bits (418), Expect = 7e-41
Identities = 113/329 (34%), Positives = 163/329 (49%), Gaps = 10/329 (3%)
Frame = +2
Query: 530 LLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIH-------VAVKVLVVGSTLT 582
L T +L AT++F +L EG FGPVY+GF+ + VAVK L +
Sbjct: 26 LYAFTLEELREATNSFSWSNMLGEGGFGPVYKGFVDDKLRSGLKAQTVAVKRLDLDGLQG 205
Query: 583 DEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDD 642
E E+ FLG+++HP+LV L GYC + R+ +Y+YM G+L+N L+ +
Sbjct: 206 HREWLAEIIFLGQLRHPHLVKLIGYCYEDEHRLLMYEYMPRGSLENQLF---RRYSAAMP 376
Query: 643 WSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSV 702
WST R KIALG A+ LAFLH P+I+R KAS++
Sbjct: 377 WST------------------------RMKIALGAAKGLAFLHE-ADKPVIYRDFKASNI 481
Query: 703 YLDYDLEPRLSDFGLAKIFGSGLDEEIA---RGSPGYVPPEFSQPEFESPTPKSDVYCFG 759
LD D +LSDFGLAK G D + G+ GY PE+ T KSDVY +G
Sbjct: 482 LLDSDFTAKLSDFGLAKDGPEGEDTHVTTRIMGTQGYAAPEYIMT--GHLTTKSDVYSYG 655
Query: 760 VVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEAL 819
VVL ELLTG++ V D + E +LV W R L+R + I + + + +
Sbjct: 656 VVLLELLTGRRVV--DKSRSNEGKSLVEWARPLLRDQKKVYNIIDRRLEGQFPMKGAMKV 829
Query: 820 KEVGYLCTADLPFKRPTMQQIVGLLKDIE 848
+ + C + P RPTM ++ +L+ ++
Sbjct: 830 AMLAFKCLSHHPNARPTMSDVIKVLEPLQ 916
>TC217385 similar to UP|Q6ZGC7 (Q6ZGC7) Receptor protein kinase PERK1-like,
partial (80%)
Length = 1604
Score = 164 bits (414), Expect = 2e-40
Identities = 109/331 (32%), Positives = 158/331 (46%), Gaps = 2/331 (0%)
Frame = +1
Query: 523 VVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLT 582
+VI + + D++ T N ++ G VY+ L VA+K +
Sbjct: 313 LVILHMNMALHVYEDIMRMTENLSEKYIIGYGASSTVYKCVLKNCKPVAIKRIYSHYPQC 492
Query: 583 DEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDD 642
+E ELE +G IKH NLV L GY ++ + YDYMENG+L +LL+ P + D
Sbjct: 493 IKEFETELETVGSIKHRNLVSLQGYSLSPYGHLLFYDYMENGSLWDLLHG-PTKKKKLD- 666
Query: 643 WSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSV 702
W R KIALG A+ LA+LHH C P IIHR VK+S++
Sbjct: 667 ------------------------WELRLKIALGAAQGLAYLHHDCCPRIIHRDVKSSNI 774
Query: 703 YLDYDLEPRLSDFGLAKIF--GSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGV 760
LD D EP L+DFG+AK G+ GY+ PE+++ T KSDVY +G+
Sbjct: 775 LLDADFEPHLTDFGIAKSLCPSKSHTSTYIMGTIGYIDPEYART--SHLTEKSDVYSYGI 948
Query: 761 VLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALK 820
VL ELLTG+K V ++ + L + N +DP I T D + +
Sbjct: 949 VLLELLTGRKAVDNE-------SNLHHLILSKAATNAVMETVDPDITATCKDLGAVKKVY 1107
Query: 821 EVGYLCTADLPFKRPTMQQIVGLLKDIEPTT 851
++ LCT P RPTM ++ +L + P++
Sbjct: 1108QLALLCTKRQPADRPTMHEVTRVLGSLVPSS 1200
>TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-like protein
, partial (47%)
Length = 1305
Score = 163 bits (412), Expect = 3e-40
Identities = 105/325 (32%), Positives = 167/325 (51%), Gaps = 5/325 (1%)
Frame = +2
Query: 531 LNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAAREL 590
L F+ + +AT F L EG FG VY+G LP VAVK L S EE E+
Sbjct: 38 LRFDFSTIEAATQKFSEANKLGEGGFGEVYKGLLPSGQEVAVKRLSKISGQGGEEFKNEV 217
Query: 591 EFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEE 650
E + +++H NLV L G+C+ G+++I +Y+++ N +L +L+D P +S D
Sbjct: 218 EIVAKLQHRNLVRLLGFCLEGEEKILVYEFVANKSLDYILFD-PEKQKSLD--------- 367
Query: 651 ADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEP 710
W R+KI G AR + +LH IIHR +KAS+V LD D+ P
Sbjct: 368 ----------------WTRRYKIVEGIARGIQYLHEDSRLKIIHRDLKASNVLLDGDMNP 499
Query: 711 RLSDFGLAKIFGSGLDEEIAR-----GSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFEL 765
++SDFG+A+IF G+D+ A G+ GY+ PE++ + KSDVY FGV++ E+
Sbjct: 500 KISDFGMARIF--GVDQTQANTNRIVGTYGYMSPEYAM--HGEYSAKSDVYSFGVLILEI 667
Query: 766 LTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYL 825
++GK+ + + A L+S+ L + +D + ++ + ++ + +G L
Sbjct: 668 ISGKR--NSSFYETDVAEDLLSYAWKLWKDEAPLELMDQSLRESYTRNEVIRCI-HIGLL 838
Query: 826 CTADLPFKRPTMQQIVGLLKDIEPT 850
C + P RPTM +V +L T
Sbjct: 839 CVQEDPIDRPTMASVVLMLDSYSVT 913
>TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis receptor
kinase 1, partial (60%)
Length = 1444
Score = 157 bits (398), Expect = 1e-38
Identities = 101/319 (31%), Positives = 162/319 (50%), Gaps = 3/319 (0%)
Frame = +1
Query: 530 LLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDE-EAAR 588
L + +L AT +F +L G FG VY+G L VAVK L T E +
Sbjct: 112 LKRFSLRELQVATDSFSNKNILGRGGFGKVYKGRLADGSLVAVKRLKEERTPGGELQFQT 291
Query: 589 ELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTW 648
E+E + H NL+ L G+C+ +R+ +Y YM NG++ + L + P Q DW T
Sbjct: 292 EVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERP-PYQEPLDWPT--- 459
Query: 649 EEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDL 708
R ++ALG+AR L++LH C P IIHR VKA+++ LD +
Sbjct: 460 ---------------------RKRVALGSARGLSYLHDHCDPKIIHRDVKAANILLDEEF 576
Query: 709 EPRLSDFGLAKI--FGSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELL 766
E + DFGLAK+ + RG+ G++ PE+ S K+DV+ +G++L EL+
Sbjct: 577 EAVVGDFGLAKLMDYKDTHVTTAVRGTIGHIAPEYLSTGKSS--EKTDVFGYGIMLLELI 750
Query: 767 TGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLC 826
TG++ + + L+ WV+GL+++ + +DP + + ++E+ L +V LC
Sbjct: 751 TGQRAFDLARLANDDDVMLLDWVKGLLKEKKLEMLVDPDLQTNYIETEVEQ-LIQVALLC 927
Query: 827 TADLPFKRPTMQQIVGLLK 845
T P RP M ++V +L+
Sbjct: 928 TQGSPMDRPKMSEVVRMLE 984
>TC218252 weakly similar to UP|Q03407 (Q03407) Ydr490cp, partial (5%)
Length = 1279
Score = 157 bits (396), Expect = 2e-38
Identities = 105/280 (37%), Positives = 147/280 (52%), Gaps = 5/280 (1%)
Frame = +1
Query: 571 AVKVLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLL 630
A+K +V + D RELE LG IKH LV L GYC + ++ IYDY+ G+L L
Sbjct: 1 ALKRIVKLNEGFDRFFERELEILGSIKHRYLVNLRGYCNSPTSKLLIYDYLPGGSLDEAL 180
Query: 631 YDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSP 690
+ E AD W R I +G A+ LA+LHH CSP
Sbjct: 181 H-----------------ERADQ-----------LDWDSRLNIIMGAAKGLAYLHHDCSP 276
Query: 691 PIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEE-----IARGSPGYVPPEFSQPE 745
IIHR +K+S++ LD +LE R+SDFGLAK+ DEE I G+ GY+ PE+ Q
Sbjct: 277 RIIHRDIKSSNILLDGNLEARVSDFGLAKLLE---DEESHITTIVAGTFGYLAPEYMQS- 444
Query: 746 FESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPK 805
T KSDVY FGV+ E+L+GK+P D ++ +V W+ L+ +N+ +DP
Sbjct: 445 -GRATEKSDVYSFGVLTLEVLSGKRPT--DAAFIEKGPNIVGWLNFLITENRPREIVDP- 612
Query: 806 ICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLK 845
+C+ G + +AL V C + P RPTM ++V LL+
Sbjct: 613 LCE-GVQMESLDALLSVAIQCVSSSPEDRPTMHRVVQLLE 729
>TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic embryogenesis
receptor-like kinase 3 {Arabidopsis thaliana;} , partial
(54%)
Length = 1008
Score = 155 bits (393), Expect = 5e-38
Identities = 104/324 (32%), Positives = 162/324 (49%), Gaps = 8/324 (2%)
Frame = +2
Query: 530 LLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTD-EEAAR 588
L + +L AT NF +L G FG VY+G L VAVK L T +
Sbjct: 110 LKRFSLRELQVATDNFSNKHILGRGGFGKVYKGRLADGSLVAVKRLKEERTQGG*LQFQT 289
Query: 589 ELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDL-----PLGVQSTDDW 643
E+E + H NL+ L G+C+ +R+ +Y YM NG++ + L + PLG
Sbjct: 290 EVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERQESQPPLG------- 448
Query: 644 STDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVY 703
W R +IALG+AR LA+LH C P IIHR VKA+++
Sbjct: 449 -----------------------WPERKRIALGSARGLAYLHDHCDPKIIHRDVKAANIL 559
Query: 704 LDYDLEPRLSDFGLAKI--FGSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVV 761
LD + E + DFGLAK+ + RG+ G++ PE+ S K+DV+ +GV+
Sbjct: 560 LDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTIGHIAPEYLSTGKSS--EKTDVFGYGVM 733
Query: 762 LFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKE 821
L EL+TG++ + + L+ WV+GL++ + +D + + +DE++E+ L +
Sbjct: 734 LLELITGQRAFDLARLANDDDVMLLDWVKGLLKDRKLETLVDADLQGSYNDEEVEQ-LIQ 910
Query: 822 VGYLCTADLPFKRPTMQQIVGLLK 845
V LCT P +RP M ++V +L+
Sbjct: 911 VALLCTQGSPMERPKMSEVVRMLE 982
>TC207146 weakly similar to UP|Q9H4D1 (Q9H4D1) Protein kinase, partial (4%)
Length = 1166
Score = 152 bits (384), Expect = 6e-37
Identities = 89/259 (34%), Positives = 137/259 (52%), Gaps = 2/259 (0%)
Frame = +1
Query: 589 ELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTW 648
E+E +GR++H +LV L GYC G R+ +Y+Y++NGNL+ L+
Sbjct: 1 EVEAIGRVRHQSLVRLLGYCAEGAHRMLVYEYVDNGNLEQWLHG---------------- 132
Query: 649 EEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDL 708
+VG LT W R I LGTA+ L +LH G P ++HR +K+S++ L
Sbjct: 133 --------DVGPCSPLT-WEIRMNIILGTAKGLTYLHEGLEPKVVHRDIKSSNILLSKKW 285
Query: 709 EPRLSDFGLAKIFGSGLDEEIAR--GSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELL 766
++SDFGLAK+ GS R G+ GYV PE++ + +SDVY FG+++ EL+
Sbjct: 286 NAKVSDFGLAKLLGSDSSYITTRVMGTFGYVAPEYASTGMLN--ERSDVYSFGILIMELI 459
Query: 767 TGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLC 826
TG+ PV DY+ E LV W++ +V +DPK+ + + ++ AL V C
Sbjct: 460 TGRNPV--DYSRPPEEVNLVDWLKKMVSNRNPEGVLDPKLPEKPTSRALKRALL-VALRC 630
Query: 827 TADLPFKRPTMQQIVGLLK 845
T KRP M ++ +L+
Sbjct: 631 TDPNAQKRPKMGHVIHMLE 687
>TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial (28%)
Length = 1358
Score = 151 bits (381), Expect = 1e-36
Identities = 101/304 (33%), Positives = 149/304 (48%), Gaps = 9/304 (2%)
Frame = +1
Query: 551 LAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVA 610
+ EG FGPVY+G +AVK L S + E E+ + ++HP+LV L G CV
Sbjct: 13 IGEGGFGPVYKGCFSDGTLIAVKQLSSKSRQGNREFLNEIGMISALQHPHLVKLYGCCVE 192
Query: 611 GDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFR 670
GDQ + +Y+YMEN +L L+ + W R
Sbjct: 193 GDQLLLVYEYMENNSLARALF-------------------------GAEEHQIKLDWTTR 297
Query: 671 HKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEE-- 728
+KI +G AR LA+LH I+HR +KA++V LD DL P++SDFGLAK LDEE
Sbjct: 298 YKICVGIARGLAYLHEESRLKIVHRDIKATNVLLDQDLNPKISDFGLAK-----LDEEDN 462
Query: 729 -----IARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEAT 783
G+ GY+ PE++ + T K+DVY FG+V E++ G+ + +E+
Sbjct: 463 THISTRIAGTFGYMAPEYAMHGY--LTDKADVYSFGIVALEIINGRS--NTIHRQKEESF 630
Query: 784 TLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALK--EVGYLCTADLPFKRPTMQQIV 841
+++ W L K +D ++ G + EEAL +V LCT RPTM +V
Sbjct: 631 SVLEWAHLLREKGDIMDLVDRRL---GLEFNKEEALVMIKVALLCTNVTAALRPTMSSVV 801
Query: 842 GLLK 845
+L+
Sbjct: 802 SMLE 813
>TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.110 precursor
- Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (31%)
Length = 1363
Score = 149 bits (375), Expect = 7e-36
Identities = 105/333 (31%), Positives = 162/333 (48%), Gaps = 4/333 (1%)
Frame = +1
Query: 516 KQATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVL 575
+Q V V +F+ +L IT +AT NF + EG FGPVY+G L G +AVK L
Sbjct: 283 RQLQDVDVPLFD--MLTIT-----AATDNFLLNNKIGEGGFGPVYKGKLVGGQEIAVKRL 441
Query: 576 VVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPL 635
S E E++ + +++H NLV L G C+ G +++ +Y+Y+ NG+L + ++D
Sbjct: 442 SSLSGQGITEFITEVKLIAKLQHRNLVKLLGCCIKGQEKLLVYEYVVNGSLNSFIFDQ-- 615
Query: 636 GVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHR 695
+ L W R I LG AR L +LH IIHR
Sbjct: 616 ------------------------IKSKLLDWPRRFNIILGIARGLLYLHQDSRLRIIHR 723
Query: 696 AVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEEIAR---GSPGYVPPEFSQPEFESP-TP 751
+KAS+V LD L P++SDFG+A+ FG E G+ GY+ PE++ F+ +
Sbjct: 724 DLKASNVLLDEKLNPKISDFGMARAFGGDQTEGNTNRVVGTYGYMAPEYA---FDGNFSI 894
Query: 752 KSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGS 811
KSDV+ FG++L E++ G K + + LV + L ++ + ID I D+
Sbjct: 895 KSDVFSFGILLLEIVCGIK--NKSLCHENQTLNLVGYAWALWKEQNALQLIDSGIKDSCV 1068
Query: 812 DEQIEEALKEVGYLCTADLPFKRPTMQQIVGLL 844
++ + V LC P RPTM ++ +L
Sbjct: 1069IPEVLRCI-HVSLLCVQQYPEDRPTMTSVIQML 1164
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,849,919
Number of Sequences: 63676
Number of extensions: 556716
Number of successful extensions: 5688
Number of sequences better than 10.0: 1044
Number of HSP's better than 10.0 without gapping: 3747
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4475
length of query: 852
length of database: 12,639,632
effective HSP length: 105
effective length of query: 747
effective length of database: 5,953,652
effective search space: 4447378044
effective search space used: 4447378044
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC122160.2


