

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121245.8 - phase: 0
(665 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
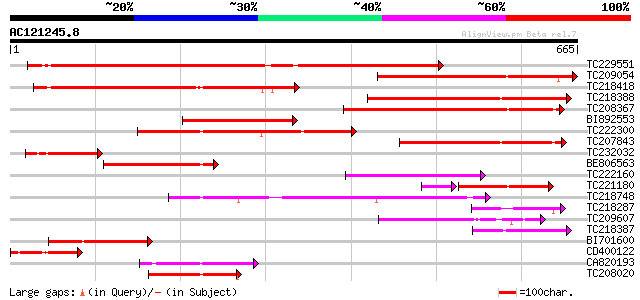
Score E
Sequences producing significant alignments: (bits) Value
TC229551 weakly similar to UP|Q6X4V5 (Q6X4V5) ABC transporter, p... 553 e-158
TC209054 similar to UP|Q6K376 (Q6K376) ABC transporter-like prot... 372 e-103
TC218418 homologue to UP|Q6X4V5 (Q6X4V5) ABC transporter, partia... 326 2e-89
TC218388 similar to UP|Q9LMU4 (Q9LMU4) F2H15.7 protein, partial ... 270 2e-72
TC208367 similar to GB|AAP37786.1|30725528|BT008427 At1g51560 {A... 226 3e-59
BI892553 216 3e-56
TC222300 similar to GB|AAP37786.1|30725528|BT008427 At1g51560 {A... 213 2e-55
TC207843 similar to GB|AAP37786.1|30725528|BT008427 At1g51560 {A... 174 9e-44
TC232032 weakly similar to UP|Q6K376 (Q6K376) ABC transporter-li... 141 1e-33
BE806563 similar to GP|11994752|db ABC transporter-like protein ... 115 6e-26
TC222160 115 6e-26
TC221180 weakly similar to UP|Q6X4V5 (Q6X4V5) ABC transporter, p... 97 1e-24
TC218748 similar to UP|Q76CU1 (Q76CU1) PDR-type ABC transporter ... 97 3e-20
TC218287 weakly similar to UP|Q6K376 (Q6K376) ABC transporter-li... 96 7e-20
TC209607 95 9e-20
TC218387 similar to UP|Q9LMU4 (Q9LMU4) F2H15.7 protein, partial ... 95 1e-19
BI701600 94 1e-19
CD400122 94 2e-19
CA820193 similar to PIR|T04229|T04 ABC-type transport protein F1... 92 1e-18
TC208020 similar to UP|Q76CU2 (Q76CU2) PDR-type ABC transporter ... 81 2e-15
>TC229551 weakly similar to UP|Q6X4V5 (Q6X4V5) ABC transporter, partial (34%)
Length = 1436
Score = 553 bits (1426), Expect = e-158
Identities = 292/492 (59%), Positives = 369/492 (74%), Gaps = 4/492 (0%)
Frame = +3
Query: 22 EEKGVCLTWKDLWVTVSASTGKTNESKSILQGLTGYAKPAQLLAIMGPSGCGKSTLLDAL 81
E+ + LTW++L TV+ GK K IL GLTGYA+P +LLAI+GPSG GKSTLLDAL
Sbjct: 3 EQNDITLTWENLEATVT--NGKNR--KLILHGLTGYAQPGRLLAIIGPSGSGKSTLLDAL 170
Query: 82 AGRLGSNTRQSGDILINGNKQALAYGTSAYVTQDDTLLTTLTVKEAVYYSAQLQLPDTMS 141
AGRL SN +Q+G ILING+KQ LAYGTS YVTQDD +L+ LT E +YYSA LQ P+TMS
Sbjct: 171 AGRLTSNIKQTGKILINGHKQELAYGTSGYVTQDDAMLSCLTAGETLYYSAMLQFPNTMS 350
Query: 142 NEEKKERADFTIREMGLQDAINTRIGGWGVKGISGGQKRRVSISIEILTRPRLLFLDEPT 201
EEKKERAD T+REMGLQDAINTR+GGW KG+SGGQ+RR+SI IEILT P+LLFLDEPT
Sbjct: 351 VEEKKERADMTLREMGLQDAINTRVGGWNCKGLSGGQRRRLSICIEILTHPKLLFLDEPT 530
Query: 202 SGLDSAASYYVMKRIASLDKKDGIQRTIVASIHQPSTEVFQLFHNLCLLSSGKTVYFGPA 261
SGLDSAASYYVM IA+L ++DGIQRTIVAS+HQPS+EVFQLFH+L LLSSG+TVYFGPA
Sbjct: 531 SGLDSAASYYVMSGIANLIQRDGIQRTIVASVHQPSSEVFQLFHDLFLLSSGETVYFGPA 710
Query: 262 SAASEFFASNGFPCPPLQNPSDHLLKTINKDFDQDIEMDLSETGTISIEQAIDILVSSYS 321
S A++FFASNGFPCPPL NPSDH L+ INKDF+QD + I+ E+A ILV+SY
Sbjct: 711 SDANQFFASNGFPCPPLYNPSDHYLRIINKDFNQDADEG------ITTEEATKILVNSYK 872
Query: 322 SSERNQEIKNEVSVLSIVLSLKDNNSTYKKK--HAGFLNQCLVLTRRSFVNMFRDLGYYW 379
SSE + +++E I S D + KKK HA F+ QCL+L RR+ + ++RD +
Sbjct: 873 SSEFSNHVQSE-----IAKSETDFGACGKKKKIHAAFITQCLILIRRASLQIYRDTNNHR 1037
Query: 380 LRLGIYIALAISLATVF-NDLDKSNGSIQDRGSLLMFVFSFLTFMT-IGGFPSYVEDMKV 437
RL ++I +++S+ ++F + SI DRGSLL F S +TFMT +GG +E+M V
Sbjct: 1038ARLVVFIFISLSVGSIFYHSGGPDLRSIMDRGSLLCFFLSVVTFMTLVGGISPLIEEMNV 1217
Query: 438 FERERLNGHYGVTAYVIGNTLSAIPYLLMISLIPGAIAYYPPGLQKGFEHFIYFICALFS 497
F+RERLNGHYG+TA++I N SA+PY ++ +IPGA+ Y GL KG ++F++ I LF+
Sbjct: 1218FKRERLNGHYGITAFLISNIFSAVPYNFLMLIIPGAVVTYLSGLHKGVDNFVFLISVLFA 1397
Query: 498 CLMLVESLMMIV 509
+ VESLMM+V
Sbjct: 1398TVTWVESLMMVV 1433
>TC209054 similar to UP|Q6K376 (Q6K376) ABC transporter-like protein, partial
(27%)
Length = 913
Score = 372 bits (956), Expect = e-103
Identities = 181/238 (76%), Positives = 211/238 (88%), Gaps = 4/238 (1%)
Frame = +1
Query: 432 VEDMKVFERERLNGHYGVTAYVIGNTLSAIPYLLMISLIPGAIAYYPPGLQKGFEHFIYF 491
VEDMKVFERERLNGHY VTA+VIGNT SAIPYLL++S+IPGAIAYY PGLQK FEHF+YF
Sbjct: 1 VEDMKVFERERLNGHYSVTAFVIGNTFSAIPYLLLVSIIPGAIAYYLPGLQKDFEHFVYF 180
Query: 492 ICALFSCLMLVESLMMIVASIVPDFLMGIITGAGIQGIMMLAGGFFRLPNDLPNPFWRYP 551
IC LF+CLMLVESLMMIVASIVP+FLMGIITGAGIQGIM++ GGFFRLPNDLP PFW+YP
Sbjct: 181 ICVLFACLMLVESLMMIVASIVPNFLMGIITGAGIQGIMIIGGGFFRLPNDLPRPFWKYP 360
Query: 552 MFYISFHRYAFQGSYKNEFEGLKFERDEIGGSLNYISGEEILRNKFHVDMSYSKWVDLGV 611
MFY++FHRYA+QG +KNEFEGL+F + +GG YISGEEILR+ + V+MSYSKW DLG+
Sbjct: 361 MFYVAFHRYAYQGLFKNEFEGLRFATNNVGG--GYISGEEILRDMWQVNMSYSKWFDLGI 534
Query: 612 LLGMIVLYRVVFLIIIKTTEKLKPIVMSFMS---VSPRQTRQILENPTATPL-HVEVV 665
LLGMI+LYRV+FLI IKTTEKLKPI++SFMS SP+ T QI++NP ATPL V+V+
Sbjct: 535 LLGMIILYRVLFLINIKTTEKLKPIIVSFMSSSRSSPKPTIQIMQNPNATPLQEVQVI 708
>TC218418 homologue to UP|Q6X4V5 (Q6X4V5) ABC transporter, partial (49%)
Length = 1220
Score = 326 bits (835), Expect = 2e-89
Identities = 177/323 (54%), Positives = 230/323 (70%), Gaps = 11/323 (3%)
Frame = +1
Query: 28 LTWKDLWVTVSASTGKTNESKSILQGLTGYAKPAQLLAIMGPSGCGKSTLLDALAGRLGS 87
LTWKDL V V+ S G+T +++L+GLTGYA+P A+MGPSG GKSTLLDAL+ RL +
Sbjct: 241 LTWKDLTVMVTLSNGET---QNVLEGLTGYAEPGTFTALMGPSGSGKSTLLDALSSRLAA 411
Query: 88 NTRQSGDILINGNKQALAYGTSAYVTQDDTLLTTLTVKEAVYYSAQLQLPDTMSNEEKKE 147
N SG IL+NG K L++GT+AYVTQDD L+ TLTV+E + YSA+L+LPD M +K+
Sbjct: 412 NAFLSGTILLNGRKAKLSFGTAAYVTQDDNLIGTLTVRETISYSARLRLPDNMPWADKRA 591
Query: 148 RADFTIREMGLQDAINTRIGGWGVKGISGGQKRRVSISIEILTRPRLLFLDEPTSGLDSA 207
+ TI MGLQD +T IG W ++GISGG+KRRVSI++EIL RPRLLFLDEPTSGLDSA
Sbjct: 592 LVESTIVAMGLQDCADTVIGNWHLRGISGGEKRRVSIALEILMRPRLLFLDEPTSGLDSA 771
Query: 208 ASYYVMKRIASLDKKDGIQRTIVASIHQPSTEVFQLFHNLCLLSSGKTVYFGPASAASEF 267
++++V + + +L +DG RT++ASIHQPS+EVF+LF L LLSSGKTVYFG AS A EF
Sbjct: 772 SAFFVTQTLRAL-ARDG--RTVIASIHQPSSEVFELFDQLYLLSSGKTVYFGQASEAYEF 942
Query: 268 FASNGFPCPPLQNPSDHLLKTINKDFDQ-----DIEMDLSETGT------ISIEQAIDIL 316
FA GFPCP L+NPSDH L+ IN DFD+ M L G+ I+ +AI L
Sbjct: 943 FAQAGFPCPALRNPSDHFLRCINSDFDKVKATLKGSMKLRFEGSDDPLDRITTAEAIRTL 1122
Query: 317 VSSYSSSERNQEIKNEVSVLSIV 339
+ Y +S+ + + +V +S V
Sbjct: 1123IDFYRTSQHSYAARQKVDEISKV 1191
>TC218388 similar to UP|Q9LMU4 (Q9LMU4) F2H15.7 protein, partial (38%)
Length = 933
Score = 270 bits (689), Expect = 2e-72
Identities = 128/240 (53%), Positives = 173/240 (71%)
Frame = +1
Query: 420 LTFMTIGGFPSYVEDMKVFERERLNGHYGVTAYVIGNTLSAIPYLLMISLIPGAIAYYPP 479
+TFM+IGGFPS+VEDMKVF+RERLNGHYGVT++VI NTLSA+P+L++I+ + G I Y+
Sbjct: 1 VTFMSIGGFPSFVEDMKVFQRERLNGHYGVTSFVISNTLSAMPFLILITFLSGTICYFMV 180
Query: 480 GLQKGFEHFIYFICALFSCLMLVESLMMIVASIVPDFLMGIITGAGIQGIMMLAGGFFRL 539
L GF H+++F+ L++ + +VESLMM +ASIVP+FLMGII GAGIQGI ML G+FRL
Sbjct: 181 RLHPGFWHYLFFVLCLYASVTVVESLMMAIASIVPNFLMGIIIGAGIQGIFMLVSGYFRL 360
Query: 540 PNDLPNPFWRYPMFYISFHRYAFQGSYKNEFEGLKFERDEIGGSLNYISGEEILRNKFHV 599
P+D+P P WRYPM YISFH +A QG Y+N+ GL F D L I GE IL F +
Sbjct: 361 PHDIPKPVWRYPMSYISFHFWALQGQYQNDLRGLVF--DNQTPDLPKIPGEYILEKVFQI 534
Query: 600 DMSYSKWVDLGVLLGMIVLYRVVFLIIIKTTEKLKPIVMSFMSVSPRQTRQILENPTATP 659
D++ SKW++L V+ MIV+YR++F I+IK E + P V +++ Q + +N T P
Sbjct: 535 DVNRSKWINLSVIFSMIVIYRIIFFIMIKVNEDVTPWVRGYLARRRMQQKSGAQNTTIAP 714
>TC208367 similar to GB|AAP37786.1|30725528|BT008427 At1g51560 {Arabidopsis
thaliana;} , partial (34%)
Length = 1062
Score = 226 bits (576), Expect = 3e-59
Identities = 111/259 (42%), Positives = 169/259 (64%)
Frame = +1
Query: 392 LATVFNDLDKSNGSIQDRGSLLMFVFSFLTFMTIGGFPSYVEDMKVFERERLNGHYGVTA 451
+ T+F + S +I RG+ F+ F+TFM+IGGFPS++E+MKVF +ERLNG+YGV
Sbjct: 1 VGTIFYGVGSSYRAIFARGACGAFISGFMTFMSIGGFPSFIEEMKVFYKERLNGYYGVGV 180
Query: 452 YVIGNTLSAIPYLLMISLIPGAIAYYPPGLQKGFEHFIYFICALFSCLMLVESLMMIVAS 511
Y++ N LS+ P++ ++S+ G I YY + F H++Y L C+ +VES MMI+AS
Sbjct: 181 YILSNFLSSFPFVAVMSIATGTITYYMVRFRTEFSHYVYICLDLIGCIAVVESSMMIIAS 360
Query: 512 IVPDFLMGIITGAGIQGIMMLAGGFFRLPNDLPNPFWRYPMFYISFHRYAFQGSYKNEFE 571
+VP+FLMG+I GAG G+MM+ G+FR DLP FWRYP+ YI++ + QG++KN+
Sbjct: 361 LVPNFLMGLIIGAGYIGVMMMTAGYFRQIPDLPKIFWRYPISYINYGAWGLQGAFKNDMI 540
Query: 572 GLKFERDEIGGSLNYISGEEILRNKFHVDMSYSKWVDLGVLLGMIVLYRVVFLIIIKTTE 631
G++F+ E GG+ + GE IL+ + + SKW DL ++ ++VL RV+F +I+K E
Sbjct: 541 GMEFDPLEPGGT--KLKGEIILKTMLGIRVEISKWWDLAAVMIILVLLRVLFFVILKFKE 714
Query: 632 KLKPIVMSFMSVSPRQTRQ 650
+ P + S+ RQT Q
Sbjct: 715 RAAPFL---YSIYARQTLQ 762
>BI892553
Length = 421
Score = 216 bits (549), Expect = 3e-56
Identities = 106/135 (78%), Positives = 118/135 (86%)
Frame = +3
Query: 203 GLDSAASYYVMKRIASLDKKDGIQRTIVASIHQPSTEVFQLFHNLCLLSSGKTVYFGPAS 262
GLDSAASYYVMKRIA+LDKKD + RT+VASIHQPS+EVFQLF NLCLLSSG+TVYFGPAS
Sbjct: 3 GLDSAASYYVMKRIATLDKKDDVHRTVVASIHQPSSEVFQLFDNLCLLSSGRTVYFGPAS 182
Query: 263 AASEFFASNGFPCPPLQNPSDHLLKTINKDFDQDIEMDLSETGTISIEQAIDILVSSYSS 322
AA EFFASNGFPCPPL NPSDHLLKTINKDFDQD E++L T TI E+AI ILV SY S
Sbjct: 183 AAKEFFASNGFPCPPLMNPSDHLLKTINKDFDQDTELNLGGTVTIPTEEAIRILVDSYKS 362
Query: 323 SERNQEIKNEVSVLS 337
SE N E++ EV+VL+
Sbjct: 363 SEMNHEVQKEVAVLT 407
>TC222300 similar to GB|AAP37786.1|30725528|BT008427 At1g51560 {Arabidopsis
thaliana;} , partial (37%)
Length = 801
Score = 213 bits (543), Expect = 2e-55
Identities = 121/269 (44%), Positives = 166/269 (60%), Gaps = 12/269 (4%)
Frame = +1
Query: 150 DFTIREMGLQDAINTRIGGWGVKGISGGQKRRVSISIEILTRPRLLFLDEPTSGLDSAAS 209
D TI EMGLQD + IG W ++GISGG+K+R+SI++ ILTRPRLLFLDEPTSGLDSA++
Sbjct: 10 DATIIEMGLQDCDDRLIGKWHLRGISGGEKKRLSIAL*ILTRPRLLFLDEPTSGLDSASA 189
Query: 210 YYVMKRIASLDKKDGIQRTIVASIHQPSTEVFQLFHNLCLLSSGKTVYFGPASAASEFFA 269
++V++ + ++ + + RT++ SIHQP +EVF L+ +L LLS G+TVYFG A +A EFF
Sbjct: 190 FFVVQTLRNVARDE---RTVIYSIHQPDSEVFALYDDLFLLSGGETVYFGEAKSAIEFFD 360
Query: 270 SNGFPCPPLQNPSDHLLKTINKDFD------------QDIEMDLSETGTISIEQAIDILV 317
GFPCP +NP DH L+ IN DFD D+ ++ + LV
Sbjct: 361 EAGFPCPRKRNPYDHFLRCINYDFDIVTATIKGSQRIHDVPNSADPFMNLATAEIKATLV 540
Query: 318 SSYSSSERNQEIKNEVSVLSIVLSLKDNNSTYKKKHAGFLNQCLVLTRRSFVNMFRDLGY 377
Y S + KN + LS L + T A + Q L LT+RSFVNM RD+GY
Sbjct: 541 EKYKRSTYARRAKNRIQELSTDEGL--HPPTQHGSQASWWKQLLTLTKRSFVNMCRDVGY 714
Query: 378 YWLRLGIYIALAISLATVFNDLDKSNGSI 406
YWLR+ +YI ++I + T + D+ S SI
Sbjct: 715 YWLRIIVYIIVSICV*TDYFDVGYSYTSI 801
>TC207843 similar to GB|AAP37786.1|30725528|BT008427 At1g51560 {Arabidopsis
thaliana;} , partial (31%)
Length = 1005
Score = 174 bits (442), Expect = 9e-44
Identities = 85/196 (43%), Positives = 131/196 (66%)
Frame = +1
Query: 458 LSAIPYLLMISLIPGAIAYYPPGLQKGFEHFIYFICALFSCLMLVESLMMIVASIVPDFL 517
LS+ P+L+ I+L I Y + G HF++F ++SC+ ++ESLMM+VAS+VP+FL
Sbjct: 1 LSSFPFLVAIALTTSTITYNMVKFRPGISHFVFFFLNIYSCISVIESLMMVVASLVPNFL 180
Query: 518 MGIITGAGIQGIMMLAGGFFRLPNDLPNPFWRYPMFYISFHRYAFQGSYKNEFEGLKFER 577
MGIITGAGI GIMM+ GFFRL +DLP P WRYP+ YIS+ +A QGSYKN+ GL+F
Sbjct: 181 MGIITGAGIIGIMMMTSGFFRLLSDLPKPVWRYPISYISYGSWAIQGSYKNDLLGLEF-- 354
Query: 578 DEIGGSLNYISGEEILRNKFHVDMSYSKWVDLGVLLGMIVLYRVVFLIIIKTTEKLKPIV 637
D + ++GE ++ + +++++SKW DL L +++ YR++F ++K E+ P+
Sbjct: 355 DPLLPGDPKLTGEYVITHMLGIELNHSKWWDLAALFVILICYRLLFFTVLKFKERASPL- 531
Query: 638 MSFMSVSPRQTRQILE 653
F ++ ++T Q LE
Sbjct: 532 --FQTLYAKRTIQQLE 573
>TC232032 weakly similar to UP|Q6K376 (Q6K376) ABC transporter-like protein,
partial (11%)
Length = 799
Score = 141 bits (355), Expect = 1e-33
Identities = 74/91 (81%), Positives = 79/91 (86%)
Frame = +2
Query: 19 NGGEEKGVCLTWKDLWVTVSASTGKTNESKSILQGLTGYAKPAQLLAIMGPSGCGKSTLL 78
N EEKG CL+WKD V V+AS GK N SKSIL+GLTGYAKP QLLAIMGPSGCGKSTLL
Sbjct: 536 NEREEKGTCLSWKD--VRVTASVGK-NGSKSILEGLTGYAKPGQLLAIMGPSGCGKSTLL 706
Query: 79 DALAGRLGSNTRQSGDILINGNKQALAYGTS 109
D LAGRLGSNTRQ+G+ILING KQALAYGTS
Sbjct: 707 DTLAGRLGSNTRQTGEILINGKKQALAYGTS 799
>BE806563 similar to GP|11994752|db ABC transporter-like protein {Arabidopsis
thaliana}, partial (20%)
Length = 428
Score = 115 bits (288), Expect = 6e-26
Identities = 62/136 (45%), Positives = 90/136 (65%), Gaps = 2/136 (1%)
Frame = +1
Query: 111 YVTQDDTLLTTLTVKEAVYYSAQLQLPDTMSNEEKKERADFTIREMGLQDAINTRIGGWG 170
+V QDD LTV E + Y+A L+LP ++S EEKKE A+ I E+GL N+ +GG
Sbjct: 28 FVPQDDVHYPHLTVLETLTYAALLRLPKSLSREEKKEHAEMVIAELGLTRCRNSPVGGCM 207
Query: 171 V--KGISGGQKRRVSISIEILTRPRLLFLDEPTSGLDSAASYYVMKRIASLDKKDGIQRT 228
+GISGG+++RVSI E+L P LLF+DEPTSGLDS + ++ + L + RT
Sbjct: 208 ALFRGISGGERKRVSIGQEMLVNPSLLFVDEPTSGLDSTTAQLIVSVLHGLARAG---RT 378
Query: 229 IVASIHQPSTEVFQLF 244
+VA+IHQPS+ ++++F
Sbjct: 379 VVATIHQPSSRLYRMF 426
>TC222160
Length = 515
Score = 115 bits (288), Expect = 6e-26
Identities = 57/165 (34%), Positives = 93/165 (55%)
Frame = +3
Query: 394 TVFNDLDKSNGSIQDRGSLLMFVFSFLTFMTIGGFPSYVEDMKVFERERLNGHYGVTAYV 453
TVF+ L S S+ R + + SF + ++I P+ ++++K++ E N H ++
Sbjct: 18 TVFSGLGHSLSSVVTRVAAIFVFVSFCSLLSIARVPALLKEIKIYACEESNQHSSTLVFL 197
Query: 454 IGNTLSAIPYLLMISLIPGAIAYYPPGLQKGFEHFIYFICALFSCLMLVESLMMIVASIV 513
+ LS+IP+L +IS+ + Y+ GL+ F +YF+ F L++ E LM+IVA++
Sbjct: 198 LAQLLSSIPFLFLISISSSLVFYFLVGLEDQFSLLMYFVLNFFMTLLVNEGLMLIVATLW 377
Query: 514 PDFLMGIITGAGIQGIMMLAGGFFRLPNDLPNPFWRYPMFYISFH 558
D ++T I MML G+FR+ N LP P W YPM YI+FH
Sbjct: 378 QDVFRSVLTLLCIHVAMMLPAGYFRVRNALPGPMWVYPMLYIAFH 512
>TC221180 weakly similar to UP|Q6X4V5 (Q6X4V5) ABC transporter, partial (8%)
Length = 948
Score = 97.1 bits (240), Expect(2) = 1e-24
Identities = 48/111 (43%), Positives = 70/111 (62%)
Frame = +2
Query: 527 QGIMMLAGGFFRLPNDLPNPFWRYPMFYISFHRYAFQGSYKNEFEGLKFERDEIGGSLNY 586
Q MM+ FR DLP FWRYPM YIS+ ++ QG +KN+ GL+FE GG+
Sbjct: 218 QVFMMVPSNIFRSLADLPKFFWRYPMSYISYITWSIQGQFKNDLIGLEFEPQVPGGT--K 391
Query: 587 ISGEEILRNKFHVDMSYSKWVDLGVLLGMIVLYRVVFLIIIKTTEKLKPIV 637
I GEEIL + F + YSKW DLGVL+ ++ YR++F +++K E++ ++
Sbjct: 392 IKGEEILHDMFGIRNDYSKWWDLGVLVLFLICYRLLFFLVLKHKERVTSLL 544
Score = 35.0 bits (79), Expect(2) = 1e-24
Identities = 16/43 (37%), Positives = 23/43 (53%), Gaps = 2/43 (4%)
Frame = +1
Query: 484 GFEHFIYFICALFSCLMLVESLMMIVASIVPDFL--MGIITGA 524
GF H+ YF +F + + E + A++ PD L MG TGA
Sbjct: 7 GFSHYCYFCMIIFCLIAVTEGCTLFFAALAPDLLVAMGTATGA 135
>TC218748 similar to UP|Q76CU1 (Q76CU1) PDR-type ABC transporter 2
(Fragment), partial (37%)
Length = 1384
Score = 96.7 bits (239), Expect = 3e-20
Identities = 88/388 (22%), Positives = 160/388 (40%), Gaps = 11/388 (2%)
Frame = +3
Query: 187 EILTRPRLLFLDEPTSGLDSAASYYVMKRIASLDKKDGIQRTIVASIHQPSTEVFQLFHN 246
E++ P ++F+DEPTSGLD+ A+ VM+ + + RT+V +IHQPS ++F+ F
Sbjct: 9 ELVANPSIIFMDEPTSGLDARAAAIVMRTVRNTVDTG---RTVVCTIHQPSIDIFESFDE 179
Query: 247 LCLL-SSGKTVYFGPASAASE-----FFASNGF-PCPPLQNPSDHLLKTINKDFDQDIEM 299
L L+ G+ +Y GP S F G NP+ +L+ + ++ +
Sbjct: 180 LLLMKQGGQEIYVGPLGHHSSHLINYFEGIQGVNKIKDGYNPATWMLEVSTSAKEMELGI 359
Query: 300 DLSETGTISIEQAIDILVSSYSSSERNQEIKNEVSVLSIVLSLKDNNSTYKKKHAGFLNQ 359
D +E Y +SE + K + LS + + FL Q
Sbjct: 360 DFAEV---------------YKNSELYRRNKALIKELSTPAPGSKDLYFPSQYSTSFLTQ 494
Query: 360 CLVLTRRSFVNMFRDLGYYWLRLGIYIALAISLATVFNDLDKSNGSIQDRGSLLMFVFSF 419
C+ + + +R+ Y +R A+A L ++F DL QD + + +++
Sbjct: 495 CMACLWKQHWSYWRNPLYTAIRFLYSTAVAAVLGSMFWDLGSKIDKQQDLFNAMGSMYAA 674
Query: 420 LTFMTIGGF----PSYVEDMKVFERERLNGHYGVTAYVIGNTLSAIPYLLMISLIPGAIA 475
+ + I P + VF RE+ G Y Y L +PY+L+ +++ G I
Sbjct: 675 VLLIGIKNANAVQPVVAVERTVFYREKAAGMYSALPYAFAQVLIELPYVLVQAVVYGIII 854
Query: 476 YYPPGLQKGFEHFIYFICALFSCLMLVESLMMIVASIVPDFLMGIITGAGIQGIMMLAGG 535
Y G + I+++ ++ + M+ ++ P+ + I + + L G
Sbjct: 855 YAMIGFEWTVTKVIWYLFFMYFTFLTFTYYGMMSVAVTPNQHISSIVSSAFYAVWNLFSG 1034
Query: 536 FFRLPNDLPNPFWRYPMFYISFHRYAFQ 563
F +P P + Y+ FQ
Sbjct: 1035FI-----VPRPVIFGSLSYLCIEICTFQ 1103
>TC218287 weakly similar to UP|Q6K376 (Q6K376) ABC transporter-like protein,
partial (5%)
Length = 775
Score = 95.5 bits (236), Expect = 7e-20
Identities = 52/119 (43%), Positives = 67/119 (55%), Gaps = 9/119 (7%)
Frame = +2
Query: 542 DLPNPFWRYPMFYISFHRYAFQGSYKNEFEGLKFERDEIGGSLNYISGEEILRNKFHVDM 601
DLP P W++P +YISF YAFQG KNEFE L F E+L + +HV M
Sbjct: 5 DLPKPVWKFPFYYISFLTYAFQGLLKNEFEDLPFS-------------SEVLADTWHVQM 145
Query: 602 SYSKWVDLGVLLGMIVLYRVVFLIIIKTTEKLKPI---------VMSFMSVSPRQTRQI 651
+SKWVDL ++ MIVLYRV+FL I K EK K + + S +++S Q R I
Sbjct: 146 GHSKWVDLAIMFAMIVLYRVLFLAISKCKEKSKQVSVGIKPETKIFSRINISELQMRGI 322
>TC209607
Length = 828
Score = 95.1 bits (235), Expect = 9e-20
Identities = 56/202 (27%), Positives = 111/202 (54%), Gaps = 6/202 (2%)
Frame = +1
Query: 433 EDMKVFERERLNGHYGVTAYVIGNTLSAIPYLLMISLIPGAIAYYPPGLQKGFEHFIYFI 492
++ ++ +E G Y V++Y I N +P+LL+++++ Y+ GL + F F++F+
Sbjct: 1 QEREILMKETSCGSYRVSSYAIANGXVYLPFLLILAILFSMPLYWLVGLNRNFLAFLHFL 180
Query: 493 CALFSCLMLVESLMMIVASIVPDFLMGIITGAGIQGIMMLAGGFFRLPNDLPNPFWRYPM 552
++ L S+++ +++VP+F++G AG+ G L G+F ++PN +W + M
Sbjct: 181 LLIWLILYTANSVVVCFSALVPNFIVGNSVIAGVIGSFFLFSGYFISKQEIPN-YWIF-M 354
Query: 553 FYISFHRYAFQGSYKNEFEGLKFERDEIGGSLNYI------SGEEILRNKFHVDMSYSKW 606
YIS +Y F+G NEF G L Y+ SGE++L+ + + S ++W
Sbjct: 355 HYISLFKYPFEGFLINEF-------SNSGKCLEYMFGACIKSGEDVLKEEGYGGES-NRW 510
Query: 607 VDLGVLLGMIVLYRVVFLIIIK 628
++GV + I++YR + +I++
Sbjct: 511 KNVGVTVCFILVYRFISYVILR 576
>TC218387 similar to UP|Q9LMU4 (Q9LMU4) F2H15.7 protein, partial (19%)
Length = 401
Score = 94.7 bits (234), Expect = 1e-19
Identities = 45/116 (38%), Positives = 67/116 (56%)
Frame = +2
Query: 544 PNPFWRYPMFYISFHRYAFQGSYKNEFEGLKFERDEIGGSLNYISGEEILRNKFHVDMSY 603
P P WRYPM YISFH +A QG Y+N+ G F+ L I G+ I F +D++
Sbjct: 2 PKPVWRYPMSYISFHFWALQGQYQNDLRG*IFDNQT--PDLPKIPGKYIWEKVFQIDVNR 175
Query: 604 SKWVDLGVLLGMIVLYRVVFLIIIKTTEKLKPIVMSFMSVSPRQTRQILENPTATP 659
SKW++L V+ MIV+YR++F I+IK E + P + +++ Q + +N T P
Sbjct: 176 SKWINLSVIFSMIVIYRIIFFIMIKVNEDVTPWIRGYLARRRMQQKSGAQNTTIAP 343
>BI701600
Length = 426
Score = 94.4 bits (233), Expect = 1e-19
Identities = 55/123 (44%), Positives = 76/123 (61%), Gaps = 1/123 (0%)
Frame = +3
Query: 46 ESKSILQGLTGYAKPAQLLAIMGPSGCGKSTLLDALAGRLGSNTRQSGDILINGNKQAL- 104
+ ++IL+G+TG A+P ++LA++GPSG GKSTLL ALAGRL +G IL N +K
Sbjct: 60 KERTILKGVTGIAQPGEILAVLGPSGSGKSTLLHALAGRL-HGPGLTGTILANSSKLTKP 236
Query: 105 AYGTSAYVTQDDTLLTTLTVKEAVYYSAQLQLPDTMSNEEKKERADFTIREMGLQDAINT 164
+ +VTQDD L LTV+E + + A L+LP + EK A+ I E+GL NT
Sbjct: 237 VLRRTGFVTQDDILYPHLTVRETLVFCAMLRLPRALLRSEKLAAAEAAIAELGLGKCENT 416
Query: 165 RIG 167
IG
Sbjct: 417 IIG 425
>CD400122
Length = 585
Score = 94.0 bits (232), Expect = 2e-19
Identities = 53/84 (63%), Positives = 60/84 (71%)
Frame = -1
Query: 2 ALEIESTREEVGASKKMNGGEEKGVCLTWKDLWVTVSASTGKTNESKSILQGLTGYAKPA 61
+ E+E + V K +E+GV LTW+DLWVTVS GK N K ILQGL GYAKP
Sbjct: 366 SFEVEKIKS-VETGKDGEEDQEEGVFLTWEDLWVTVS--NGK-NGRKPILQGLKGYAKPG 199
Query: 62 QLLAIMGPSGCGKSTLLDALAGRL 85
+LLAIMGPSGCGKSTLLDALAG L
Sbjct: 198 KLLAIMGPSGCGKSTLLDALAGNL 127
>CA820193 similar to PIR|T04229|T04 ABC-type transport protein F14M19.30 -
Arabidopsis thaliana, partial (23%)
Length = 423
Score = 91.7 bits (226), Expect = 1e-18
Identities = 51/139 (36%), Positives = 80/139 (56%)
Frame = +1
Query: 153 IREMGLQDAINTRIGGWGVKGISGGQKRRVSISIEILTRPRLLFLDEPTSGLDSAASYYV 212
+ E+ L NTR+ +G+SGG++RRVSI + +L P +L LDEPTSGLDS +++ V
Sbjct: 25 LSELRLTHLSNTRLA----RGLSGGERRRVSIGLCLLHDPAVLLLDEPTSGLDSTSAFKV 192
Query: 213 MKRIASLDKKDGIQRTIVASIHQPSTEVFQLFHNLCLLSSGKTVYFGPASAASEFFASNG 272
M+ + RTI+ SIHQPS ++ + LLS G+ V+ G + F SNG
Sbjct: 193 MRILKQTCVSR--NRTIILSIHQPSFKILACIDRILLLSKGQVVHHGSVATLQAFLHSNG 366
Query: 273 FPCPPLQNPSDHLLKTINK 291
F P N ++ ++ +++
Sbjct: 367 FTVPHQLNALEYAMEILSQ 423
>TC208020 similar to UP|Q76CU2 (Q76CU2) PDR-type ABC transporter 1, partial
(14%)
Length = 765
Score = 80.9 bits (198), Expect = 2e-15
Identities = 43/115 (37%), Positives = 72/115 (62%), Gaps = 5/115 (4%)
Frame = +2
Query: 163 NTRIGGWGVKGISGGQKRRVSISIEILTRPRLLFLDEPTSGLDSAASYYVMKRIASLDKK 222
N+ +G GV G+S Q++R++I++E++ P ++F+DEPTSGLD+ A+ VM+ + +
Sbjct: 206 NSLVGLPGVSGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMRTVRNTVDT 385
Query: 223 DGIQRTIVASIHQPSTEVFQLFHNLCLLS-SGKTVYFGPASAAS----EFFASNG 272
RT+V +IHQPS ++F+ F L L+ G+ +Y GP S ++F S G
Sbjct: 386 G---RTVVCTIHQPSIDIFEAFDELFLMKRGGQEIYVGPLGRHSTHLIKYFESIG 541
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.138 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,478,003
Number of Sequences: 63676
Number of extensions: 349930
Number of successful extensions: 2232
Number of sequences better than 10.0: 127
Number of HSP's better than 10.0 without gapping: 2165
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2190
length of query: 665
length of database: 12,639,632
effective HSP length: 103
effective length of query: 562
effective length of database: 6,081,004
effective search space: 3417524248
effective search space used: 3417524248
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC121245.8


