

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121245.12 + phase: 0 /pseudo
(136 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
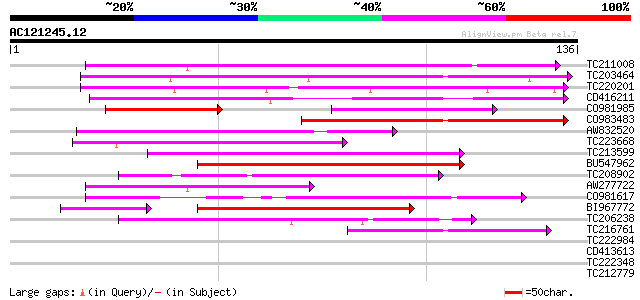
Score E
Sequences producing significant alignments: (bits) Value
TC211008 64 2e-11
TC203464 similar to GB|AAF35920.1|7025820|AC005892 L505.8 {Leish... 62 1e-10
TC220201 similar to UP|Q9ERI5 (Q9ERI5) Apoptotic cell clearance ... 57 3e-09
CD416211 55 1e-08
CO981985 40 2e-08
CO983483 51 1e-07
AW832520 50 3e-07
TC223668 47 2e-06
TC213599 homologue to UP|EXBB_PASHA (P72202) Biopolymer transpor... 47 3e-06
BU547962 45 8e-06
TC208902 43 5e-05
AW277722 similar to SP|O65719|HS73_ Heat shock cognate 70 kDa pr... 41 2e-04
CO981617 40 3e-04
BI967772 37 5e-04
TC206238 weakly similar to UP|Q7PX96 (Q7PX96) AgCP12237 (Fragmen... 39 8e-04
TC216761 similar to UP|Q9FQE4 (Q9FQE4) Glutathione S-transferase... 39 8e-04
TC222984 39 0.001
CD413613 38 0.001
TC222348 34 0.024
TC212779 33 0.042
>TC211008
Length = 516
Score = 64.3 bits (155), Expect = 2e-11
Identities = 40/118 (33%), Positives = 64/118 (53%), Gaps = 4/118 (3%)
Frame = +1
Query: 19 WNPPPEDFIKINVDGSSFGNPDN----AGFGGLLRNNRGNWIHGFSGSCGRATNLFAELS 74
W P ++ +N DGS F N N + GGL+R++ G ++ GF+ + G + AEL
Sbjct: 64 WRCPXNGWVCLNTDGSVFENHRNGCSGSACGGLVRDSSGCYLGGFTVNLGNTSVTLAELW 243
Query: 75 AIWKGLQLAWDLGYRSIIMESDSQVALDLILDTKQKDFHPHASLLSLIRKLSSLPWVV 132
+ GL+LAWDLG + + ++ DS AL L+ + P +L+S I +L W+V
Sbjct: 244 GVVHGLKLAWDLGCKKVKVDIDSGNALGLVRHGPVAN-DPAFALVSEINELVRKEWLV 414
>TC203464 similar to GB|AAF35920.1|7025820|AC005892 L505.8 {Leishmania
major;} , partial (8%)
Length = 1386
Score = 61.6 bits (148), Expect = 1e-10
Identities = 40/121 (33%), Positives = 62/121 (51%), Gaps = 3/121 (2%)
Frame = +2
Query: 18 RWNPPPEDFIKINVDGSSFG-NPDNAGFGGLLRNNRGNWIHGFSGSCGRATNLF-AELSA 75
RW P ++K+NVDGS +AG GG+LR+ W+ GF+ + EL A
Sbjct: 485 RWKKPEIGWVKLNVDGSRDPYKSSSAGCGGVLRDASAKWLRGFAKKLNPTYAVHQTELEA 664
Query: 76 IWKGLQLAWDLGYRSIIMESDSQVALDLILDTKQKDFHPHASLLSLIR-KLSSLPWVVYL 134
I GL++A ++ + +I+ESDS + ++ + K HP ++ LIR K W V
Sbjct: 665 ILTGLKVASEMNVKKLIVESDSDSVVSMV-ENGVKPNHPDYGVVELIRTKRRRFDWEVRF 841
Query: 135 V 135
V
Sbjct: 842 V 844
>TC220201 similar to UP|Q9ERI5 (Q9ERI5) Apoptotic cell clearance receptor
PtdSerR (Phosphatidylserine receptor), partial (5%)
Length = 1185
Score = 57.0 bits (136), Expect = 3e-09
Identities = 39/127 (30%), Positives = 66/127 (51%), Gaps = 10/127 (7%)
Frame = +1
Query: 18 RWNPPPEDFIKINVDGSSFGN-PDNAGFGGLLRNNRGNWIHGFS-----GSCGRATNLFA 71
+W P ++K+NVDGS P +AG GG++R+ G W GF C +A +
Sbjct: 334 KWKKPESGWVKLNVDGSRIHEEPASAGCGGVIRDEWGTWCVGFDQKLDPNICRQAH--YT 507
Query: 72 ELSAIWKGLQLAWD--LGYRSIIMESDSQVALDLILDTKQKDFH-PHASLLSLIRKLSSL 128
EL AI GL++A + + +++ESDS+ A++++ +H P ++ I +L
Sbjct: 508 ELQAILTGLKVAREDMINVEKLVVESDSEPAVNMVKSRLGYKYHRPEYKVVQDINRLLKD 687
Query: 129 P-WVVYL 134
P W+ L
Sbjct: 688 PKWLARL 708
>CD416211
Length = 551
Score = 55.1 bits (131), Expect = 1e-08
Identities = 41/123 (33%), Positives = 57/123 (46%), Gaps = 8/123 (6%)
Frame = -3
Query: 20 NPPPEDFIKINVDGSSFGNPDNAGFGGLLRNNRGNWIHGFSG--------SCGRATNLFA 71
+PP + IKIN DGSSFGNP +G+GG+ RN G+ F G CG +N
Sbjct: 477 SPPDPNTIKINTDGSSFGNPGISGYGGVFRNEHGHVASWFLGQQWVYD*HKCGVVSN--- 307
Query: 72 ELSAIWKGLQLAWDLGYRSIIMESDSQVALDLILDTKQKDFHPHASLLSLIRKLSSLPWV 131
+ G +S+I ESDS ++L ++ + P S+ LS P V
Sbjct: 306 -----------SKQKG*QSVICESDSALSLLMV-------YSPSCSVSWRYSWLSFCPVV 181
Query: 132 VYL 134
YL
Sbjct: 180 CYL 172
>CO981985
Length = 903
Score = 40.4 bits (93), Expect(2) = 2e-08
Identities = 16/28 (57%), Positives = 23/28 (82%)
Frame = -2
Query: 24 EDFIKINVDGSSFGNPDNAGFGGLLRNN 51
++ +K+NVDGSS GNP GFGG+LR++
Sbjct: 482 DNAVKLNVDGSSLGNPS*GGFGGILRDS 399
Score = 33.5 bits (75), Expect(2) = 2e-08
Identities = 17/40 (42%), Positives = 24/40 (59%)
Frame = -1
Query: 78 KGLQLAWDLGYRSIIMESDSQVALDLILDTKQKDFHPHAS 117
+GL LAW Y++I +SDS +AL L++ H HAS
Sbjct: 372 RGLTLAWSAVYKNIECDSDSLLALQLLV*EGVTIHHQHAS 253
>CO983483
Length = 829
Score = 51.2 bits (121), Expect = 1e-07
Identities = 29/64 (45%), Positives = 39/64 (60%)
Frame = +2
Query: 71 AELSAIWKGLQLAWDLGYRSIIMESDSQVALDLILDTKQKDFHPHASLLSLIRKLSSLPW 130
AE A++ GL++A D G RS+I ESDS++AL LI FHPHA L+ I+ + W
Sbjct: 578 AEQFALYHGLKVAEDKGIRSLICESDSKMALQLI-QHNDNAFHPHALLIRKIQSYKNWQW 754
Query: 131 VVYL 134
V L
Sbjct: 755 EVQL 766
>AW832520
Length = 423
Score = 50.4 bits (119), Expect = 3e-07
Identities = 24/77 (31%), Positives = 40/77 (51%)
Frame = -3
Query: 17 VRWNPPPEDFIKINVDGSSFGNPDNAGFGGLLRNNRGNWIHGFSGSCGRATNLFAELSAI 76
+RW+PPPE K+N DG+ N GG++R++ GN++ GF+ + T AEL
Sbjct: 256 IRWSPPPEGLFKLNYDGAVNCNSLKVSAGGVVRDSHGNFVIGFTTTPDSCTINIAEL--- 86
Query: 77 WKGLQLAWDLGYRSIIM 93
W W + +++
Sbjct: 85 WASTD*EWPISMDFVVL 35
>TC223668
Length = 866
Score = 47.4 bits (111), Expect = 2e-06
Identities = 24/68 (35%), Positives = 36/68 (52%), Gaps = 2/68 (2%)
Frame = -3
Query: 16 QVRWNPPPE--DFIKINVDGSSFGNPDNAGFGGLLRNNRGNWIHGFSGSCGRATNLFAEL 73
++ W PP + +K+NVDGS P GGL+R+ G W+ F+ CG L +EL
Sbjct: 594 RLTWTSPPPLLNEVKLNVDGSGHYEPHVMEVGGLIRDGVGRWLFRFARHCGLGNPLLSEL 415
Query: 74 SAIWKGLQ 81
A+ L+
Sbjct: 414 FALEASLR 391
>TC213599 homologue to UP|EXBB_PASHA (P72202) Biopolymer transport exbB
protein, partial (8%)
Length = 825
Score = 47.0 bits (110), Expect = 3e-06
Identities = 28/76 (36%), Positives = 38/76 (49%)
Frame = +3
Query: 34 SSFGNPDNAGFGGLLRNNRGNWIHGFSGSCGRATNLFAELSAIWKGLQLAWDLGYRSIIM 93
S + D A GGLLR+ G W F G + AEL + GL LAWD G +++ +
Sbjct: 327 SMHSDNDMAACGGLLRDCHGRWAASFV*KLGNTSAFVAEL*GAYLGLSLAWDKGDKNLEV 506
Query: 94 ESDSQVALDLILDTKQ 109
DS V + I +KQ
Sbjct: 507 NIDSLVVVVSIKISKQ 554
>BU547962
Length = 591
Score = 45.4 bits (106), Expect = 8e-06
Identities = 19/64 (29%), Positives = 42/64 (64%)
Frame = -2
Query: 46 GLLRNNRGNWIHGFSGSCGRATNLFAELSAIWKGLQLAWDLGYRSIIMESDSQVALDLIL 105
G++RN+ ++ ++ S G+A + AEL+A+ KGL+L + G+ I +E D++ +++I+
Sbjct: 590 GVVRNHNAEFLXXYAESIGQANSTIAELTALRKGLELVLENGWNDIWLEGDAKTLVEIIV 411
Query: 106 DTKQ 109
++
Sbjct: 410 KRRK 399
>TC208902
Length = 838
Score = 42.7 bits (99), Expect = 5e-05
Identities = 27/78 (34%), Positives = 45/78 (57%)
Frame = +1
Query: 27 IKINVDGSSFGNPDNAGFGGLLRNNRGNWIHGFSGSCGRATNLFAELSAIWKGLQLAWDL 86
I N DG+ G A G +LRN G + F+ + G + AE+ AI G++LAWD
Sbjct: 232 INSNCDGAVSGGV--AACGRVLRNQAGAVVV-FAHNLGMCSISQAEIWAITMGVKLAWDK 402
Query: 87 GYRSIIMESDSQVALDLI 104
G+ ++ +ESD + A++++
Sbjct: 403 GFTNLYVESDFKYAIEMM 456
>AW277722 similar to SP|O65719|HS73_ Heat shock cognate 70 kDa protein 3
(Hsc70.3). [Mouse-ear cress] {Arabidopsis thaliana},
partial (8%)
Length = 432
Score = 40.8 bits (94), Expect = 2e-04
Identities = 21/59 (35%), Positives = 33/59 (55%), Gaps = 4/59 (6%)
Frame = +1
Query: 19 WNPPPEDFIKINVDGSSFGNPDN----AGFGGLLRNNRGNWIHGFSGSCGRATNLFAEL 73
W PP ++ +N DGS F N N + GGL+R++ G ++ GF+ + G + AEL
Sbjct: 247 WRCPPNGWVCLNTDGSVFENHRNGCSGSACGGLVRDSSGCYLGGFTVNLGNTSVTLAEL 423
>CO981617
Length = 837
Score = 40.4 bits (93), Expect = 3e-04
Identities = 33/106 (31%), Positives = 52/106 (48%)
Frame = -1
Query: 19 WNPPPEDFIKINVDGSSFGNPDNAGFGGLLRNNRGNWIHGFSGSCGRATNLFAELSAIWK 78
W+PP K+N DG F + W+ SC + L A+L +I+
Sbjct: 351 WSPPCPGVSKVNCDGVVF-----------IHEAIATWM----ASC---SILEAKLWSIFY 226
Query: 79 GLQLAWDLGYRSIIMESDSQVALDLILDTKQKDFHPHASLLSLIRK 124
G++LA D G + IIMESDS A++ +++ D HP L+ I++
Sbjct: 225 GMRLAEDCGVQHIIMESDSVEAVNHLME-HSYDNHPLFHLVQEIKR 91
>BI967772
Length = 719
Score = 37.4 bits (85), Expect(2) = 5e-04
Identities = 19/52 (36%), Positives = 32/52 (61%)
Frame = +3
Query: 46 GLLRNNRGNWIHGFSGSCGRATNLFAELSAIWKGLQLAWDLGYRSIIMESDS 97
GL R++ G+ + F+ L AE I+ GL +AW+ G++++I+ESDS
Sbjct: 435 GLQRDHSGDVLFCFAAKVDYCFVLHAESWTIYIGLIIAWEEGFKNLIIESDS 590
Score = 21.2 bits (43), Expect(2) = 5e-04
Identities = 7/22 (31%), Positives = 13/22 (58%)
Frame = +1
Query: 13 VICQVRWNPPPEDFIKINVDGS 34
+I ++ WN P +K+N + S
Sbjct: 340 LISRICWNKAPTGVLKLNCNAS 405
>TC206238 weakly similar to UP|Q7PX96 (Q7PX96) AgCP12237 (Fragment), partial
(9%)
Length = 1259
Score = 38.9 bits (89), Expect = 8e-04
Identities = 26/92 (28%), Positives = 47/92 (50%), Gaps = 6/92 (6%)
Frame = +3
Query: 27 IKINVDGSSFGNPDNAGFGGLLRNNRGNWIHGFSGSCGRA-TNLFAELSAIWKGLQLA-- 83
+++NV G+ + D A GG+ R+N ++ GFS G +N E+ I+ G+++A
Sbjct: 624 LRLNVSGAYDRSSDTAACGGIFRDNNDRFVLGFSVKLGECLSNDEGEIWGIYHGMKIARR 803
Query: 84 ---WDLGYRSIIMESDSQVALDLILDTKQKDF 112
W L YR I+ + ++ + +Q DF
Sbjct: 804 YDIWGL-YRDILRQHSIPISAKI---ARQHDF 887
>TC216761 similar to UP|Q9FQE4 (Q9FQE4) Glutathione S-transferase GST 14
(Fragment) , complete
Length = 1353
Score = 38.9 bits (89), Expect = 8e-04
Identities = 21/49 (42%), Positives = 28/49 (56%)
Frame = +1
Query: 82 LAWDLGYRSIIMESDSQVALDLILDTKQKDFHPHASLLSLIRKLSSLPW 130
+AW G R++I ESDS +AL LI T HP A+L++ IR W
Sbjct: 1018 IAWTRGIRNLICESDSSMALQLI-STGVYITHPRAALVTTIRSFMDKDW 1161
>TC222984
Length = 970
Score = 38.5 bits (88), Expect = 0.001
Identities = 18/42 (42%), Positives = 26/42 (61%)
Frame = -2
Query: 7 PTNKGHVICQVRWNPPPEDFIKINVDGSSFGNPDNAGFGGLL 48
P+ ++ V W+P ED +K+ DGSSF NPD +G G +L
Sbjct: 177 PSKPQRLVTLVSWHPC-EDVVKLKTDGSSFRNPDPSG*GDIL 55
>CD413613
Length = 623
Score = 38.1 bits (87), Expect = 0.001
Identities = 27/90 (30%), Positives = 44/90 (48%), Gaps = 3/90 (3%)
Frame = -1
Query: 18 RWNPPPEDFIKINVDGSSFGNPDNAGFGGLLRNNRGNWIHGFSGSC---GRATNLFAELS 74
+W+ P +FIK N+D S F N G G RN++G ++ SC G + AE
Sbjct: 485 KWSRPSLEFIKCNLDASLFRNYLLHGLGACNRNSQGTFVADL--SCWFKGLLPPMEAEDR 312
Query: 75 AIWKGLQLAWDLGYRSIIMESDSQVALDLI 104
A+ + + + + +I+E D Q +D I
Sbjct: 311 ALSQVITWLLNNNFTKVILEIDCQQLVDTI 222
>TC222348
Length = 995
Score = 33.9 bits (76), Expect = 0.024
Identities = 22/80 (27%), Positives = 39/80 (48%)
Frame = -2
Query: 22 PPEDFIKINVDGSSFGNPDNAGFGGLLRNNRGNWIHGFSGSCGRATNLFAELSAIWKGLQ 81
P +IK N D ++ GNP AG G L + ++ F+ S + FAE+ A ++
Sbjct: 328 PDYPWIKCNTDCAARGNPGLAGCGKLFIYSNARFLGNFAYSIAINSAFFAEVMAAIIAIE 149
Query: 82 LAWDLGYRSIIMESDSQVAL 101
A + + + +++DS L
Sbjct: 148 KAVENRWNFLWLKTDSSFQL 89
>TC212779
Length = 617
Score = 33.1 bits (74), Expect = 0.042
Identities = 19/57 (33%), Positives = 28/57 (48%), Gaps = 6/57 (10%)
Frame = +1
Query: 3 YALGPTNKGHV-----ICQVRWNPPPEDFIKINVDGSSFGNPDN-AGFGGLLRNNRG 53
Y + P +K +V + V W PPP ++K NV+ F GFG LR+ +G
Sbjct: 208 YGMEPDSKVYVCSAELVISVVWQPPPTSYLKCNVNVVFFNLQHKIGGFGMCLRDEKG 378
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.140 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,965,537
Number of Sequences: 63676
Number of extensions: 111122
Number of successful extensions: 571
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 564
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 566
length of query: 136
length of database: 12,639,632
effective HSP length: 87
effective length of query: 49
effective length of database: 7,099,820
effective search space: 347891180
effective search space used: 347891180
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC121245.12


