

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121241.13 - phase: 0 /pseudo
(869 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
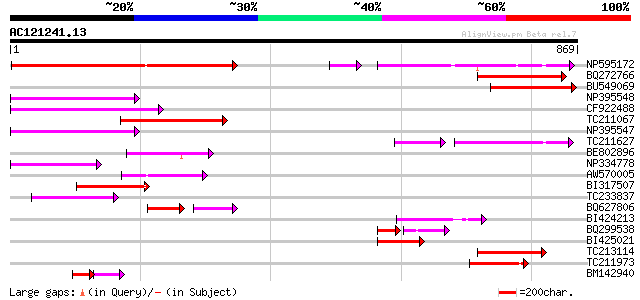
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 339 4e-93
BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis ... 178 1e-44
BU549069 177 2e-44
NP395548 reverse transcriptase [Glycine max] 149 5e-36
CF922488 145 1e-34
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 144 2e-34
NP395547 reverse transcriptase [Glycine max] 138 9e-33
TC211627 70 4e-26
BE802896 104 1e-22
NP334778 reverse transcriptase [Glycine max] 100 4e-21
AW570005 98 1e-20
BI317507 97 2e-20
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 91 2e-18
BQ627806 51 2e-14
BI424213 77 3e-14
BQ299538 51 2e-13
BI425021 70 4e-12
TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, parti... 69 9e-12
TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 67 4e-11
BM142940 44 9e-10
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 339 bits (869), Expect = 4e-93
Identities = 168/346 (48%), Positives = 237/346 (67%)
Frame = +1
Query: 3 DYRQLNKVTIKNRYPLPRIDDLMDQLVGARVFSKIDLSSGYHQIKVKDEDMQKTAFRTRY 62
DYR LN +T+K+ +P+P +D+L+D+L GA+ FSK+DL SGYHQI V+ ED +KTAFRT +
Sbjct: 1954 DYRALNAITVKDSFPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQPEDREKTAFRTHH 2133
Query: 63 GHYEYKVMPFGVTNAPGVFMEYMNRIFHAYLDKFVVVFSDDILIYSKNEEEHAEHMKIVL 122
GHYE+ VMPFG+TNAP F MN+IF L KFV+VF DDILIYS + ++H +H++ VL
Sbjct: 2134 GHYEWLVMPFGLTNAPATFQCLMNKIFQFALRKFVLVFFDDILIYSASWKDHLKHLESVL 2313
Query: 123 QLLKEKKLYAKLSKCEFWLSEVSFLGHIISGSGIVVDPSKVDAVSQWETPKSVTEIRSFL 182
Q LK+ +L+A+LSKC F +EV +LGH +SG G+ ++ +KV AV W TP +V ++R FL
Sbjct: 2314 QTLKQHQLFARLSKCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVKQLRGFL 2493
Query: 183 GLAGYYRRFIEGFSKLALPLTQLTCKGKSFVWDAQCENSFNELKRRLTTAPILILPKPDE 242
GL GYYRRFI+ ++ +A PLT L K SF+W+ + E +F +LK+ +T AP+L LP +
Sbjct: 2494 GLTGYYRRFIKSYANIAGPLTDLLQK-DSFLWNNEAEAAFVKLKKAMTEAPVLSLPDFSQ 2670
Query: 243 PFVVYCDASKLGLGGVLMQDGKVVAYASRQLRIHEKNYPTHDLELAAVVFVLKIWRHYLY 302
PF++ DAS +G+G VL Q+G +AY S++L + + EL A+ L +RHYL
Sbjct: 2671 PFILETDASGIGVGAVLGQNGHPIAYFSKKLAPRMQKQSAYTRELLAITEALSKFRHYLL 2850
Query: 303 GS*FEVFSDHKSLKYLFDQKELNMRQRRWLELLKDCDFGLNYHPGK 348
G+ F + +D +SLK L DQ Q+ WL DF + Y PGK
Sbjct: 2851 GNKFIIRTDQRSLKSLMDQSLQTPEQQAWLHKFLGYDFKIEYKPGK 2988
Score = 158 bits (399), Expect(2) = 4e-39
Identities = 105/309 (33%), Positives = 157/309 (49%), Gaps = 8/309 (2%)
Frame = +1
Query: 565 LAEIYIHNIVKLHGVPSSIVSDRNFRFTSRFWKSLQDALGSKLRVSSAYHPQADGHSERT 624
+AE ++ +IVKLHG+P SIVSDR+ FTS FW+ L G+ L +SSAYHPQ+DG SE
Sbjct: 3562 VAEAFMSHIVKLHGIPRSIVSDRDRVFTSTFWQHLFKLQGTTLAMSSAYHPQSDGQSEVL 3741
Query: 625 IQSLEDLLRVCVLEQGGAWDSHLPLIEFTYNNSYHSSIGMAPFETLYGRRCRTPLCWFES 684
+ LE LR E W LP EF YN +YH S+GM PF LYGR T
Sbjct: 3742 NKCLEMYLRCFTYEHPKGWVKALPWAEFWYNTAYHMSLGMTPFRALYGREPPT----LTR 3909
Query: 685 GESVLLGPDLVHQTIEKVQMIREKMKASQSR----QKSYHDKRKKALEFQEGDHVFLRVT 740
+ P V + + + K+K + +R K DK++ + FQ GD V +++
Sbjct: 3910 QACSIDDPAEVREQLTDRDALLAKLKINLTRAQQVMKRQADKKRLDVSFQIGDEVLVKLQ 4089
Query: 741 PMTGVGRAL-KSKKLTPKFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKY--- 796
P L K++KL+ ++ GP+++ ++G VAY++ L P + +H VFHVSQL+ +
Sbjct: 4090 PYRQHSAVLRKNQKLSMRYFGPFKVLAKIGDVAYKLEL-PSAARIHPVFHVSQLKPFNGT 4266
Query: 797 VADPSHVIPRADVQVRDNLTIETLPLRIEYREVKKLRGKDIPLVKVVLGGVTGESLTWEL 856
DP +P ++ + P++I + I + V + TWE
Sbjct: 4267 AQDPYLPLPLTVTEMGPVMQ----PVKILASRIIIRGHNQIEQILVQWENGLQDEATWED 4434
Query: 857 ESKMLESYP 865
+ SYP
Sbjct: 4435 IEDIKASYP 4461
Score = 22.7 bits (47), Expect(2) = 4e-39
Identities = 15/49 (30%), Positives = 21/49 (42%)
Frame = +3
Query: 491 CCSVCLCMFDLSEV*G*ASEACRVVDTIRCARVEVGQHFHGFCDEFAEY 539
C S+ M DLS * A R T + +G+ HGF F ++
Sbjct: 3324 CQSLYTEMLDLSTS*IKQYTASRTTPTFTYSSTSLGRCSHGFHHRFTKF 3470
>BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis melo},
partial (9%)
Length = 410
Score = 178 bits (451), Expect = 1e-44
Identities = 88/136 (64%), Positives = 108/136 (78%)
Frame = -3
Query: 718 SYHDKRKKALEFQEGDHVFLRVTPMTGVGRALKSKKLTPKFIGPYQISERVGTVAYRVGL 777
SYHDKR+K LEF+ GDHVFLRVTP TGVGRALKS KLTP FIGP+QI ++V VAY++ L
Sbjct: 408 SYHDKRRKDLEFEVGDHVFLRVTP*TGVGRALKS*KLTPHFIGPFQILKKVDFVAYQIAL 229
Query: 778 PPHLSNLHDVFHVSQLRKYVADPSHVIPRADVQVRDNLTIETLPLRIEYREVKKLRGKDI 837
PP L++LH+VFHVSQ+ KY+ DPSH++ VQV++NL ETLPLRI+ K LRGK+I
Sbjct: 228 PPSLTSLHNVFHVSQIHKYINDPSHMVNLDVVQVKENLAHETLPLRIKDMRTKHLRGKEI 49
Query: 838 PLVKVVLGGVTGESLT 853
LVKV+ GG +GE T
Sbjct: 48 LLVKVIWGGASGEEAT 1
>BU549069
Length = 615
Score = 177 bits (449), Expect = 2e-44
Identities = 87/132 (65%), Positives = 104/132 (77%)
Frame = -1
Query: 737 LRVTPMTGVGRALKSKKLTPKFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKY 796
L+VTP TGVG+ALKS+KLTP FIG +QI +R VAY++ LPP LSNLH+VFHVSQLR Y
Sbjct: 615 LKVTPRTGVGQALKSRKLTPHFIGHFQILKRAXPVAYQIALPPSLSNLHNVFHVSQLRMY 436
Query: 797 VADPSHVIPRADVQVRDNLTIETLPLRIEYREVKKLRGKDIPLVKVVLGGVTGESLTWEL 856
+ DPSHV+ DVQV++NLT ETLPLRIE R K LR K+ PLVKV+ GG +GE TWEL
Sbjct: 435 IHDPSHVVKLDDVQVKENLTYETLPLRIEDRRTKHLRRKENPLVKVIWGGTSGEDATWEL 256
Query: 857 ESKMLESYPELF 868
ES+M +YP LF
Sbjct: 255 ESQMRVAYPSLF 220
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 149 bits (376), Expect = 5e-36
Identities = 78/197 (39%), Positives = 117/197 (58%)
Frame = +1
Query: 2 IDYRQLNKVTIKNRYPLPRIDDLMDQLVGARVFSKIDLSSGYHQIKVKDEDMQKTAFRTR 61
IDYR+LN+ T K+ +PLP +D ++++L G + +D GY+QI V +D +K AF
Sbjct: 172 IDYRKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCP 351
Query: 62 YGHYEYKVMPFGVTNAPGVFMEYMNRIFHAYLDKFVVVFSDDILIYSKNEEEHAEHMKIV 121
+G + Y+ +PFG+ NAP F M IF ++K + VF DD ++ + E + +++V
Sbjct: 352 FGVFAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMV 531
Query: 122 LQLLKEKKLYAKLSKCEFWLSEVSFLGHIISGSGIVVDPSKVDAVSQWETPKSVTEIRSF 181
LQ E L KC F + E LGH IS GI VD +K+D + + P +V IRSF
Sbjct: 532 LQRCVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSF 711
Query: 182 LGLAGYYRRFIEGFSKL 198
LG A +YRRFI+ F+K+
Sbjct: 712 LGQARFYRRFIKDFTKV 762
>CF922488
Length = 741
Score = 145 bits (365), Expect = 1e-34
Identities = 80/235 (34%), Positives = 126/235 (53%)
Frame = +3
Query: 2 IDYRQLNKVTIKNRYPLPRIDDLMDQLVGARVFSKIDLSSGYHQIKVKDEDMQKTAFRTR 61
+DYR LN + K+++PLP I+ L+D FS +D SGY+QIK+ EDM+KT F T
Sbjct: 36 VDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAPEDMEKTTFITL 215
Query: 62 YGHYEYKVMPFGVTNAPGVFMEYMNRIFHAYLDKFVVVFSDDILIYSKNEEEHAEHMKIV 121
+G + YK M FG+ N + M +F + K + V+ DD+++ S+ EEEH +++ +
Sbjct: 216 WGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRTEEEHLVNLRKL 395
Query: 122 LQLLKEKKLYAKLSKCEFWLSEVSFLGHIISGSGIVVDPSKVDAVSQWETPKSVTEIRSF 181
+ L++ +L +KC F + L I S GI VD +KV + + P + +++ F
Sbjct: 396 FRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMAKPHTEKQVQGF 575
Query: 182 LGLAGYYRRFIEGFSKLALPLTQLTCKGKSFVWDAQCENSFNELKRRLTTAPILI 236
LG Y RFI PL L CK + WD C +F +K+ L +L+
Sbjct: 576 LGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLINPHVLV 740
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 144 bits (362), Expect = 2e-34
Identities = 73/163 (44%), Positives = 102/163 (61%)
Frame = +1
Query: 171 TPKSVTEIRSFLGLAGYYRRFIEGFSKLALPLTQLTCKGKSFVWDAQCENSFNELKRRLT 230
T KSV +IRSF GLA +YRRF+ FS +A PL +L K +F W + E +F LK +LT
Sbjct: 100 TLKSVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKLT 279
Query: 231 TAPILILPKPDEPFVVYCDASKLGLGGVLMQDGKVVAYASRQLRIHEKNYPTHDLELAAV 290
AP+L LP + F + CDAS +G+ VL+Q G +AY S +L NYPT+D EL A+
Sbjct: 280 KAPVLALPDFSKTFELECDASGVGVRAVLLQGGHPIAYFSEKLHSATLNYPTYDKELYAL 459
Query: 291 VFVLKIWRHYLYGS*FEVFSDHKSLKYLFDQKELNMRQRRWLE 333
+ + W H+L F + SDH+SLKY+ + +LN R +W+E
Sbjct: 460 IRAPQTWEHFLVCKEFVIHSDHQSLKYIRGKSKLNKRHAKWVE 588
Score = 38.9 bits (89), Expect = 0.010
Identities = 22/76 (28%), Positives = 38/76 (49%)
Frame = +2
Query: 139 FWLSEVSFLGHIISGSGIVVDPSKVDAVSQWETPKSVTEIRSFLGLAGYYRRFIEGFSKL 198
F + + F G ++ +G+ +DP K+ A+ +W P V EI LG + + IEG +
Sbjct: 2 FCVDNIFFSGFVVGRNGVQMDPEKIKAIQEWPPP*KVWEI---LGASMG*QASIEGSFLI 172
Query: 199 ALPLTQLTCKGKSFVW 214
+L L L+ +W
Sbjct: 173 SLQLHHLSMSW*RRIW 220
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 138 bits (348), Expect = 9e-33
Identities = 75/197 (38%), Positives = 110/197 (55%)
Frame = +1
Query: 2 IDYRQLNKVTIKNRYPLPRIDDLMDQLVGARVFSKIDLSSGYHQIKVKDEDMQKTAFRTR 61
IDYR+LN+ T K+ YPLP +D ++ +L + +D SGY+QI V +D +KTAF
Sbjct: 172 IDYRKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCP 351
Query: 62 YGHYEYKVMPFGVTNAPGVFMEYMNRIFHAYLDKFVVVFSDDILIYSKNEEEHAEHMKIV 121
+ + Y+ MPFG+ NA F M IF ++K + VF DD + + +++ V
Sbjct: 352 FSVFAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKV 531
Query: 122 LQLLKEKKLYAKLSKCEFWLSEVSFLGHIISGSGIVVDPSKVDAVSQWETPKSVTEIRSF 181
LQ ++ L KC F + E LGH IS GI V K+D + + P +V I SF
Sbjct: 532 LQRCEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSF 711
Query: 182 LGLAGYYRRFIEGFSKL 198
LG G+YRRFI+ F+K+
Sbjct: 712 LGHVGFYRRFIKDFTKV 762
>TC211627
Length = 1034
Score = 70.5 bits (171), Expect(2) = 4e-26
Identities = 50/185 (27%), Positives = 88/185 (47%), Gaps = 2/185 (1%)
Frame = +3
Query: 682 FESGESVLLGPDLVHQTIEKVQ-MIREKMKASQSRQKSYHDKRKKALEFQEGDHVFLRVT 740
+ +G S + D + ++ + + +++ Q K D ++ L F GD V++R+
Sbjct: 276 YSAGTSTVEAVDAILHSLATIHHTLTCRLQKYQDSMKRIADSHRRDLTFNIGDWVYVRL* 455
Query: 741 PMTGVGRALKSKKLTPKFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVAD- 799
P KL+ +F GPYQI RVG VAYR+ LPP S +H +FHVS L+ +
Sbjct: 456 PYRQTSIQSTYTKLSKRFYGPYQIQARVGQVAYRLQLPP-TSKIHPIFHVSLLKVHHGPI 632
Query: 800 PSHVIPRADVQVRDNLTIETLPLRIEYREVKKLRGKDIPLVKVVLGGVTGESLTWELESK 859
P ++ ++ ++ PL+ ++ + IP V V + E TWE ++
Sbjct: 633 PPELLALPPFSTTNHPLVQ--PLQFLDWKMDESTTPPIPQVLVQWTNLAPEDTTWESWTQ 806
Query: 860 MLESY 864
+ + Y
Sbjct: 807 LKDIY 821
Score = 67.0 bits (162), Expect(2) = 4e-26
Identities = 36/78 (46%), Positives = 42/78 (53%)
Frame = +2
Query: 591 FTSRFWKSLQDALGSKLRVSSAYHPQADGHSERTIQSLEDLLRVCVLEQGGAWDSHLPLI 650
F S W L G+KLR S+AYHPQ DG +E + LE LR V + W L L
Sbjct: 5 FISGLWHELFHISGTKLRFSTAYHPQTDGQTEVINRILEQYLRAFVHDHPQHWFKFLSLA 184
Query: 651 EFTYNNSYHSSIGMAPFE 668
E YN S HS IG +PFE
Sbjct: 185 E*CYNTSVHSGIGFSPFE 238
>BE802896
Length = 416
Score = 104 bits (260), Expect = 1e-22
Identities = 61/137 (44%), Positives = 82/137 (59%), Gaps = 4/137 (2%)
Frame = -2
Query: 180 SFLGLAGYYRRFIEGFSKLALPLTQLTCKGKSFVWDAQCENSFNELKRRLTTAPILILPK 239
SFLG AG+YRRFI F K+ALPL+ L K F ++ +C+ +F+ LKR L T PI+ P
Sbjct: 415 SFLGHAGFYRRFIRDFRKVALPLSNLLQKEVEFDFNDKCK*AFDCLKRALITTPIIQAPD 236
Query: 240 PDEPFVVYCDASKLGLGGVLMQD----GKVVAYASRQLRIHEKNYPTHDLELAAVVFVLK 295
PF + CDAS LG VL Q +V+ Y+SR L + NY T + EL A+VF L+
Sbjct: 235 WTAPFELMCDASNYALGVVLAQKIDKLPRVIYYSSRTLDAAQANYTTTEKELLAIVFALE 56
Query: 296 IWRHYLYGS*FEVFSDH 312
+ YL G+ V+ DH
Sbjct: 55 KFHSYLLGTRIIVYIDH 5
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 100 bits (248), Expect = 4e-21
Identities = 52/140 (37%), Positives = 82/140 (58%)
Frame = +3
Query: 2 IDYRQLNKVTIKNRYPLPRIDDLMDQLVGARVFSKIDLSSGYHQIKVKDEDMQKTAFRTR 61
+DYR LN+ + K+ +PLP ID LM + +FS +D SGY+QIK+ EDM+KT F T
Sbjct: 12 VDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTFITL 191
Query: 62 YGHYEYKVMPFGVTNAPGVFMEYMNRIFHAYLDKFVVVFSDDILIYSKNEEEHAEHMKIV 121
+G + YKVM FG+ N + M +F + K + + D+++ S+ EEEH +++ +
Sbjct: 192 WGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNLQNL 371
Query: 122 LQLLKEKKLYAKLSKCEFWL 141
L++ +L KC F L
Sbjct: 372 FGQLRKYRLRLNPRKCVFGL 431
>AW570005
Length = 413
Score = 98.2 bits (243), Expect = 1e-20
Identities = 53/132 (40%), Positives = 77/132 (58%)
Frame = -2
Query: 172 PKSVTEIRSFLGLAGYYRRFIEGFSKLALPLTQLTCKGKSFVWDAQCENSFNELKRRLTT 231
P++ +R FL L G+YRRFI+G++ +A PL+ L K SFVW + + +F LK +T
Sbjct: 406 PRTARSLRGFLRLTGFYRRFIKGYAAMAAPLSHLLTKD-SFVWSPEADVAFQALKNVVTN 230
Query: 232 APILILPKPDEPFVVYCDASKLGLGGVLMQDGKVVAYASRQLRIHEKNYPTHDLELAAVV 291
+L LP +PF V DAS +G VL Q+G +A+ S++ T+ ELAA+
Sbjct: 229 TLVLALPDFTKPFTVETDASGSDMGAVLSQEGHPIAFFSKEFCPKLVRSSTYVHELAAIT 50
Query: 292 FVLKIWRHYLYG 303
V+K WR YL G
Sbjct: 49 NVVKKWRQYLLG 14
>BI317507
Length = 359
Score = 97.4 bits (241), Expect = 2e-20
Identities = 50/113 (44%), Positives = 72/113 (63%), Gaps = 1/113 (0%)
Frame = -1
Query: 103 DILIYSKNEEEHAEHMKIVLQLLKEKKLYAKLSKCEFWLSEVSFLGHIISGSGIVVDPSK 162
+ILIYS + + H H+ VL +LK+++L A KC F + + +LGH+IS + +D +K
Sbjct: 353 NILIYSPDWKSHIMHLTAVLDVLKKERLVANRKKCYFSQTTIEYLGHVISKDCVAMDSNK 174
Query: 163 VDAVSQWETPKSVTEIRSFLGLAGYYRRFIEGFSKLA-LPLTQLTCKGKSFVW 214
V +V +W PK+V + SFL L GYYR+FI+ + KLA PLT LT K F W
Sbjct: 173 VKSVIEWPVPKNVKRVCSFLRLTGYYRKFIKDYGKLAPRPLTDLT-KNDGFKW 18
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 91.3 bits (225), Expect = 2e-18
Identities = 48/133 (36%), Positives = 76/133 (57%)
Frame = +2
Query: 34 FSKIDLSSGYHQIKVKDEDMQKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAYL 93
FS +D SGY+QI + ED++KT F T +G + Y+VM FG+ N + M +FH +
Sbjct: 2 FSFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMM 181
Query: 94 DKFVVVFSDDILIYSKNEEEHAEHMKIVLQLLKEKKLYAKLSKCEFWLSEVSFLGHIISG 153
K + V+ DD++ S+ E EH ++ + L++ +L +KC F + LG I+S
Sbjct: 182 HKEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQ 361
Query: 154 SGIVVDPSKVDAV 166
GI +DP KV A+
Sbjct: 362 KGIEIDPEKVKAL 400
>BQ627806
Length = 435
Score = 50.8 bits (120), Expect(2) = 2e-14
Identities = 26/67 (38%), Positives = 36/67 (52%)
Frame = +1
Query: 282 THDLELAAVVFVLKIWRHYLYGS*FEVFSDHKSLKYLFDQKELNMRQRRWLELLKDCDFG 341
T+ ELAA+ +K WR YL G F + +DH+SLK L Q Q+ +L L D+
Sbjct: 232 TYVRELAAITVAVKKWRQYLLGHHFVILTDHRSLKELMSQAVQTPEQQIYLARLMGFDYT 411
Query: 342 LNYHPGK 348
+ Y GK
Sbjct: 412 IQYRAGK 432
Score = 47.0 bits (110), Expect(2) = 2e-14
Identities = 22/57 (38%), Positives = 36/57 (62%)
Frame = +3
Query: 212 FVWDAQCENSFNELKRRLTTAPILILPKPDEPFVVYCDASKLGLGGVLMQDGKVVAY 268
F W+ + + +F++LK L AP+L LP + FVV DAS +G+G +L Q+ +A+
Sbjct: 21 FHWNEEADRAFSQLKLALCQAPVLGLPDFNSSFVVETDASGIGMGAILSQNHHPLAF 191
>BI424213
Length = 426
Score = 77.0 bits (188), Expect = 3e-14
Identities = 45/138 (32%), Positives = 68/138 (48%)
Frame = +1
Query: 593 SRFWKSLQDALGSKLRVSSAYHPQADGHSERTIQSLEDLLRVCVLEQGGAWDSHLPLIEF 652
S FWK+L LG+KL S+ HPQ DG ++ +SL LLR + +WD +LP +EF
Sbjct: 4 SHFWKTLWAKLGTKLLFSTTCHPQTDGQTKVVNRSLSTLLRALLKGNHKSWDEYLPHVEF 183
Query: 653 TYNNSYHSSIGMAPFETLYGRRCRTPLCWFESGESVLLGPDLVHQTIEKVQMIREKMKAS 712
YN H + +PFE +YG TPL DL+ ++ I ++ ++
Sbjct: 184 AYNRGVHRTTKQSPFEVVYGFNPLTPL-------------DLIPLPLD-TSFIHKEGESR 321
Query: 713 QSRQKSYHDKRKKALEFQ 730
K H++ K +E Q
Sbjct: 322 SEFVKKMHERVKNQIENQ 375
>BQ299538
Length = 426
Score = 51.2 bits (121), Expect(2) = 2e-13
Identities = 27/71 (38%), Positives = 40/71 (56%)
Frame = +3
Query: 604 GSKLRVSSAYHPQADGHSERTIQSLEDLLRVCVLEQGGAWDSHLPLIEFTYNNSYHSSIG 663
G+ L++S++YHP DG + LE LR V +Q L E+ YN ++H+S G
Sbjct: 144 GTYLKMSTSYHP*IDGQTVN--HCLETFLRCFVADQPKM*VQWLSWAEYWYNTNFHASTG 317
Query: 664 MAPFETLYGRR 674
PFE +YGR+
Sbjct: 318 TTPFEVVYGRK 350
Score = 43.5 bits (101), Expect(2) = 2e-13
Identities = 19/35 (54%), Positives = 25/35 (71%)
Frame = +1
Query: 565 LAEIYIHNIVKLHGVPSSIVSDRNFRFTSRFWKSL 599
LAEI+ +V LHGVP+S++SD + F S FWK L
Sbjct: 28 LAEIFTKEVVHLHGVPASVLSDEDPIFVSSFWKEL 132
>BI425021
Length = 426
Score = 70.1 bits (170), Expect = 4e-12
Identities = 34/72 (47%), Positives = 48/72 (66%)
Frame = -1
Query: 565 LAEIYIHNIVKLHGVPSSIVSDRNFRFTSRFWKSLQDALGSKLRVSSAYHPQADGHSERT 624
+A ++++ ++KLHG P SIVSDR+ F S FW+ L G+ LR+SSAYHPQ DG +E
Sbjct: 261 VASLFLNIVIKLHGFPRSIVSDRDPLFISHFWQDLFRLSGTVLRMSSAYHPQTDGQTEVL 82
Query: 625 IQSLEDLLRVCV 636
+ +E LR V
Sbjct: 81 NRVIEQYLRAFV 46
>TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, partial (7%)
Length = 810
Score = 68.9 bits (167), Expect = 9e-12
Identities = 39/108 (36%), Positives = 66/108 (61%), Gaps = 1/108 (0%)
Frame = +3
Query: 717 KSYHDKRKKALEFQEGDHVFLRVTPMTGVGRALK-SKKLTPKFIGPYQISERVGTVAYRV 775
K+Y D+ ++A+ GD V+L++ P A K ++KL+P+F GPYQI +++G VA+ +
Sbjct: 6 KAYADRSRRAVTLSVGDWVYLKLQPYRLKSLAKKRNEKLSPRFYGPYQIKKQIGLVAFEL 185
Query: 776 GLPPHLSNLHDVFHVSQLRKYVADPSHVIPRADVQVRDNLTIETLPLR 823
LPP +H VFH S L+K VA ++ P + + ++L+ E L+
Sbjct: 186 DLPP-ARKIHPVFHASLLKKAVAATANPQP-LPLMLSEDLSSEFFQLK 323
>TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 730
Score = 66.6 bits (161), Expect = 4e-11
Identities = 35/92 (38%), Positives = 59/92 (64%), Gaps = 1/92 (1%)
Frame = +1
Query: 705 IREKMKASQSRQKSYHDKRKKALEFQEGDHVFLRVTPMTGVGRALK-SKKLTPKFIGPYQ 763
+RE + S + +KR++ +E+ GD VFL++ P A + ++KL+P+F P+Q
Sbjct: 64 LRENLLKS*DIM*ANTNKRRRDIEYVVGD*VFLKMQPYRRRSLAKRINEKLSPRFYAPFQ 243
Query: 764 ISERVGTVAYRVGLPPHLSNLHDVFHVSQLRK 795
+ +VGT+AY++ LP H+ +H VFHVS L+K
Sbjct: 244 VFNKVGTIAYKLDLPSHI-KIHPVFHVSLLKK 336
>BM142940
Length = 422
Score = 43.5 bits (101), Expect(2) = 9e-10
Identities = 21/34 (61%), Positives = 25/34 (72%)
Frame = -1
Query: 97 VVVFSDDILIYSKNEEEHAEHMKIVLQLLKEKKL 130
+ FS DILIYS NE +H +H+KIVLQ KE KL
Sbjct: 260 ICFFSYDILIYSINEGKHEKHLKIVLQTFKEAKL 159
Score = 38.5 bits (88), Expect(2) = 9e-10
Identities = 16/47 (34%), Positives = 25/47 (53%)
Frame = -3
Query: 129 KLYAKLSKCEFWLSEVSFLGHIISGSGIVVDPSKVDAVSQWETPKSV 175
K+ A KC F + +L H+IS G+ DP K++ + W PK +
Sbjct: 165 KVGANRKKCSFGCKKFEYLVHMISVKGVSADPKKLEVMVAWPIPKEM 25
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.337 0.148 0.477
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,458,317
Number of Sequences: 63676
Number of extensions: 606793
Number of successful extensions: 4054
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 3998
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4041
length of query: 869
length of database: 12,639,632
effective HSP length: 106
effective length of query: 763
effective length of database: 5,889,976
effective search space: 4494051688
effective search space used: 4494051688
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC121241.13


