

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
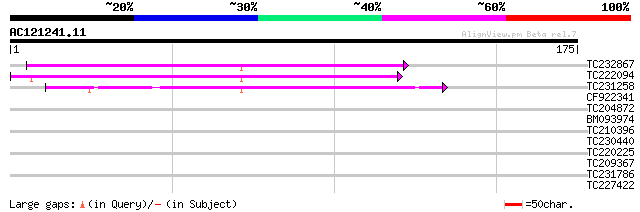
Query= AC121241.11 + phase: 0 /pseudo
(175 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC232867 similar to UP|Q9MA68 (Q9MA68) F2J6.13 protein, partial ... 87 3e-18
TC222094 84 3e-17
TC231258 similar to UP|Q886T6 (Q886T6) Sensory box histidine kin... 62 1e-10
CF922341 33 0.053
TC204872 homologue to UP|Q84LL5 (Q84LL5) Cullin 4, partial (42%) 27 4.9
BM093974 homologue to GP|7303964|gb|A CG8181-PA {Drosophila mela... 27 4.9
TC210396 similar to UP|Q75ZN6 (Q75ZN6) RNA-directed RNA Polymera... 27 6.4
TC230440 similar to UP|Q9FML2 (Q9FML2) Histone deacetylase, part... 27 6.4
TC220225 similar to UP|SY61_ARATH (Q946Y7) Syntaxin 61 (AtSYP61)... 27 6.4
TC209367 similar to UP|Q9LH58 (Q9LH58) GTP-binding protein-like,... 26 8.4
TC231786 similar to UP|HS30_ONCTS (P42931) Heat shock protein 30... 26 8.4
TC227422 similar to UP|P92973 (P92973) CCA1 (MYB-related transcr... 26 8.4
>TC232867 similar to UP|Q9MA68 (Q9MA68) F2J6.13 protein, partial (3%)
Length = 1144
Score = 87.4 bits (215), Expect = 3e-18
Identities = 43/121 (35%), Positives = 69/121 (56%), Gaps = 3/121 (2%)
Frame = -3
Query: 6 ALKEYAIISSDEPYDAILYPTVEGNNFEIKSALIYLVQENQFFGSPTEDPNLHVSIFLRL 65
AL++Y+ + + + +I P V+ +LI L+Q N F G P EDP H++ ++ +
Sbjct: 905 ALEDYSSSTVPQFFTSIAQPEVQAQTITYPPSLIQLIQSNFFHGLPNEDPYAHLATYIEI 726
Query: 66 SGTLK---ANQEAVRLHLFPFSLRDRASAWFHSLKIGSIISWDQMRKAFLSQFFPHSKTA 122
T++ +A+RL L FSL A W HS K S+ SWD++ + FL ++FP SKTA
Sbjct: 725 CNTIRLAGVPADAIRLSLLSFSLSGEAKRWLHSFKGNSLKSWDEVVEKFLKKYFPESKTA 546
Query: 123 Q 123
+
Sbjct: 545 E 543
>TC222094
Length = 984
Score = 84.3 bits (207), Expect = 3e-17
Identities = 45/128 (35%), Positives = 72/128 (56%), Gaps = 7/128 (5%)
Frame = -2
Query: 1 MAEKH----ALKEYAIISSDEPYDAILYPTVEGNNFEIKSALIYLVQENQFFGSPTEDPN 56
MAE H L++Y+ + + +I P V+ NF +LI L+Q N F+G PTEDP
Sbjct: 872 MAENHPQRMTLEDYSSPIIPQYFTSIARPEVQTANFSYPYSLIQLIQGNLFYGLPTEDPY 693
Query: 57 LHVSIFLRLSGTLK---ANQEAVRLHLFPFSLRDRASAWFHSLKIGSIISWDQMRKAFLS 113
H++ ++ + T+K ++A+ L LF FSL A W S K ++ +W++ + FL
Sbjct: 692 AHLATYIDICNTVKIVGVPEDAIHLDLFCFSLAGEARTWLRSFKGNNLRTWNEXXEKFLK 513
Query: 114 QFFPHSKT 121
++FP SKT
Sbjct: 512 KYFPESKT 489
>TC231258 similar to UP|Q886T6 (Q886T6) Sensory box histidine kinase/response
regulator, partial (3%)
Length = 1374
Score = 62.4 bits (150), Expect = 1e-10
Identities = 40/129 (31%), Positives = 71/129 (55%), Gaps = 5/129 (3%)
Frame = +2
Query: 12 IISSDEPYDAIL--YPTVEGNNFEIKSALIYLVQENQFFGSPTEDPNLHVSIFLRLSGTL 69
+++ D Y+++ YP E + +K+ LI+L+ + F G EDP+ H+ F + T+
Sbjct: 452 MVAPDFTYESLCNQYPD-EDVPYVLKTGLIHLLPK--FHGLVGEDPHKHLKEFHIVCSTM 622
Query: 70 K---ANQEAVRLHLFPFSLRDRASAWFHSLKIGSIISWDQMRKAFLSQFFPHSKTAQLRI 126
K ++ + L FP SL A W + L SI +WD +++ FL +FFP S+T +R
Sbjct: 623 KPPDVQEDHIFLKAFPHSLEGVAKDWLYYLAPRSIFNWDDLKRVFLEKFFPASRTTTIR- 799
Query: 127 KSHSSLKKM 135
K S ++++
Sbjct: 800 KDISGIRQL 826
>CF922341
Length = 675
Score = 33.5 bits (75), Expect = 0.053
Identities = 18/64 (28%), Positives = 31/64 (48%)
Frame = +1
Query: 52 TEDPNLHVSIFLRLSGTLKANQEAVRLHLFPFSLRDRASAWFHSLKIGSIISWDQMRKAF 111
T P H+ ++ + G A E + +H F SL A W+ +L+ + SW + AF
Sbjct: 448 TTCPKNHLKMYCQKMGAY-AKDEELLIHSFQESLTGVAVTWYTNLEPSRVHSWKDLMVAF 624
Query: 112 LSQF 115
+ Q+
Sbjct: 625 VRQY 636
>TC204872 homologue to UP|Q84LL5 (Q84LL5) Cullin 4, partial (42%)
Length = 1224
Score = 26.9 bits (58), Expect = 4.9
Identities = 13/54 (24%), Positives = 28/54 (51%)
Frame = +1
Query: 13 ISSDEPYDAILYPTVEGNNFEIKSALIYLVQENQFFGSPTEDPNLHVSIFLRLS 66
+S ++ + I +P ++GN I + L+ FF DPN + +I+++ +
Sbjct: 1027 VSMNDSFFDIFHPFIDGNCTVIIKHVYSLLSFTIFFQLEVTDPNPYTNIWIKFN 1188
>BM093974 homologue to GP|7303964|gb|A CG8181-PA {Drosophila melanogaster},
partial (2%)
Length = 429
Score = 26.9 bits (58), Expect = 4.9
Identities = 19/58 (32%), Positives = 29/58 (49%), Gaps = 2/58 (3%)
Frame = +1
Query: 83 FSLRDRASAWFHSLKIGSIISWDQMRKAFLSQFFPHSK--TAQLRIKSHSSLKKMVNL 138
++L ASAW + SW +AF + PH K +A R K+ S+K+MV +
Sbjct: 79 YTLHAMASAW--------VKSWQCKSRAFEDVYHPHPKILSASCR-KTMESIKEMVEI 225
>TC210396 similar to UP|Q75ZN6 (Q75ZN6) RNA-directed RNA Polymerase
(Fragment), partial (82%)
Length = 919
Score = 26.6 bits (57), Expect = 6.4
Identities = 16/57 (28%), Positives = 30/57 (52%)
Frame = +3
Query: 47 FFGSPTEDPNLHVSIFLRLSGTLKANQEAVRLHLFPFSLRDRASAWFHSLKIGSIIS 103
++G TE L +I +++S + ++A +++ SLR A AWF+ G + S
Sbjct: 360 YYGIKTEAEILGGNI-MKMSKSFNKRRDAEAINMAVRSLRKEARAWFNENSSGDVDS 527
>TC230440 similar to UP|Q9FML2 (Q9FML2) Histone deacetylase, partial (45%)
Length = 1032
Score = 26.6 bits (57), Expect = 6.4
Identities = 19/42 (45%), Positives = 21/42 (49%)
Frame = -2
Query: 64 RLSGTLKANQEAVRLHLFPFSLRDRASAWFHSLKIGSIISWD 105
R+S TLK N +RL L D A SL I SII WD
Sbjct: 209 RVSSTLKNN--CIRLINIHDFLNDGAKQTTESLIIQSIIEWD 90
>TC220225 similar to UP|SY61_ARATH (Q946Y7) Syntaxin 61 (AtSYP61) (Osmotic
stess-sensitive mutant 1), partial (76%)
Length = 1204
Score = 26.6 bits (57), Expect = 6.4
Identities = 19/46 (41%), Positives = 27/46 (58%), Gaps = 5/46 (10%)
Frame = +3
Query: 113 SQFFPHSKTAQLRIKSHSSL--KKMVNL---FTMRGSVSKKCLGFA 153
S+ F +SKT LR+K HS+L KK NL + +R + K+ L A
Sbjct: 48 SEAFRNSKTQCLRLKIHSTLSSKKFKNLLISYNLRFTNGKRLLMLA 185
>TC209367 similar to UP|Q9LH58 (Q9LH58) GTP-binding protein-like, partial
(35%)
Length = 995
Score = 26.2 bits (56), Expect = 8.4
Identities = 17/43 (39%), Positives = 22/43 (50%), Gaps = 1/43 (2%)
Frame = -3
Query: 43 QENQFFGSPTEDPNLHVS-IFLRLSGTLKANQEAVRLHLFPFS 84
Q NQ+F DP + ++ IF L + V LHLFPFS
Sbjct: 699 QNNQYFHEKMSDPMV*LTPIFDHLM*DFLGSLLQVCLHLFPFS 571
>TC231786 similar to UP|HS30_ONCTS (P42931) Heat shock protein 30 (HSP 30),
partial (7%)
Length = 1171
Score = 26.2 bits (56), Expect = 8.4
Identities = 13/44 (29%), Positives = 23/44 (51%)
Frame = -1
Query: 5 HALKEYAIISSDEPYDAILYPTVEGNNFEIKSALIYLVQENQFF 48
H L+ ++ PY + YP G+N ++S + +Q N+FF
Sbjct: 688 HHLRLVRHLACHTPYTLLKYPRTMGSNCFLQSCSL-SIQNNEFF 560
>TC227422 similar to UP|P92973 (P92973) CCA1 (MYB-related transcription
factor (CCA1); supported by cDNA: gi:1777442) (MYB
transcription factor), partial (11%)
Length = 901
Score = 26.2 bits (56), Expect = 8.4
Identities = 18/60 (30%), Positives = 28/60 (46%), Gaps = 4/60 (6%)
Frame = -2
Query: 72 NQEAVRLHLFPFSLRDRASAWFHSLKIGSIISWDQM----RKAFLSQFFPHSKTAQLRIK 127
N++ +LH FPF L+ H S+ W + + F+S FF + QLRI+
Sbjct: 360 NKQITKLHTFPFFLK-------H*NPQNSVQKWTSLATKQKHCFVSLFFYPTLLFQLRIE 202
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.138 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,901,874
Number of Sequences: 63676
Number of extensions: 104742
Number of successful extensions: 622
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 619
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 621
length of query: 175
length of database: 12,639,632
effective HSP length: 91
effective length of query: 84
effective length of database: 6,845,116
effective search space: 574989744
effective search space used: 574989744
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC121241.11


