

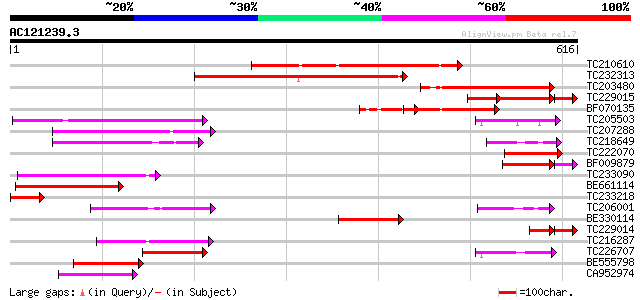
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121239.3 + phase: 0 /pseudo
(616 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC210610 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, par... 384 e-107
TC232313 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, par... 312 2e-85
TC203480 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, par... 207 1e-53
TC229015 similar to UP|Q8GT52 (Q8GT52) Hexose transporter, parti... 119 4e-46
BF070135 137 1e-32
TC205503 UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, complete 128 9e-30
TC207288 UP|Q7XA51 (Q7XA51) Monosaccharide transporter, complete 94 2e-19
TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein... 91 1e-18
TC222070 weakly similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_1... 87 3e-17
BF009879 weakly similar to GP|26986186|em hexose transporter {Ho... 80 2e-16
TC233090 similar to UP|Q9ZVN7 (Q9ZVN7) T7A14.10 protein, partial... 83 3e-16
BE661114 83 4e-16
TC233218 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, par... 75 9e-14
TC206001 similar to UP|Q8VZI5 (Q8VZI5) AT5g18840/F17K4_90, parti... 72 9e-13
BE330114 similar to PIR|T45780|T457 sugar transporter-like prote... 71 1e-12
TC229014 similar to UP|Q8GT52 (Q8GT52) Hexose transporter, parti... 48 3e-12
TC216287 similar to UP|Q8GTR0 (Q8GTR0) Sugar transporter, partia... 65 9e-11
TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter... 60 4e-09
BE555798 homologue to GP|1353516|gb|A sugar transporter {Medicag... 57 2e-08
CA952974 similar to GP|17381265|gb| AT5g18840/F17K4_90 {Arabidop... 56 5e-08
>TC210610 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, partial (27%)
Length = 674
Score = 384 bits (985), Expect = e-107
Identities = 193/230 (83%), Positives = 205/230 (88%)
Frame = +3
Query: 263 EHGQSWVARPVTGQSSVGLVSRKGSMANPSGLVDPLVTLFGSVHEKLPETGSMRSTLFPH 322
E GQSWVARPV G +SVGLVSRKGSMANPS LVDPLVTLFGSVHEKLPETGS TLFPH
Sbjct: 3 EQGQSWVARPVAGPNSVGLVSRKGSMANPSSLVDPLVTLFGSVHEKLPETGS---TLFPH 173
Query: 323 FGSMFSVGGNQPRNEDWDEESLAREGDDYISDAAAGDSDDNLQSPLISRQTTSMDKDMPL 382
FGSMFSVGGNQPRNEDWDEESLAREGDDY+SDA GDSDDNLQSPLISRQTTSMDKD+
Sbjct: 174 FGSMFSVGGNQPRNEDWDEESLAREGDDYVSDA--GDSDDNLQSPLISRQTTSMDKDITP 347
Query: 383 PAQGSLSNMRQGSLLQGNAGEPVGSTGIGGGWQLAWKWSEQEGPGGKKEGGFKRIYLHQE 442
A +L++MRQGSLL GNAGEP GSTGIGGGWQLAWKWSE+E P GKKEGGFKRIYLHQ+
Sbjct: 348 HAHSNLASMRQGSLLHGNAGEPTGSTGIGGGWQLAWKWSERESPDGKKEGGFKRIYLHQD 527
Query: 443 GGPGSIRASVVSLPGGDVPTDGDVVQQAAALVSQPALYNKELMHQQPVGP 492
GG GS R SVVSLPGGD+PTD +VV QAAALVSQPALYNK+LM Q+PV P
Sbjct: 528 GGSGSRRGSVVSLPGGDLPTDSEVV-QAAALVSQPALYNKDLMRQRPVRP 674
>TC232313 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, partial (23%)
Length = 716
Score = 312 bits (800), Expect = 2e-85
Identities = 162/239 (67%), Positives = 195/239 (80%), Gaps = 7/239 (2%)
Frame = +1
Query: 201 AKKVLQRLRGCQDVAGEMALLVEGLGVGGDTSIEEYIIGPDNELADEEDPSTGKDQIKLY 260
AKKVLQRLRG +DV+GEMALLVEGLG+GGDTSIEEYIIGP +++AD + +T KD+I+LY
Sbjct: 1 AKKVLQRLRGREDVSGEMALLVEGLGIGGDTSIEEYIIGPADKVADGHEHATEKDKIRLY 180
Query: 261 GPEHGQSWVARPVTGQSSVGLVSRKGSMANPS-GLVDPLVTLFGSVHEKLPET---GSMR 316
G + G SW+A+PVTGQSS+GL SR GS+ N S L+DPLVTLFGS+HEKLPET GSMR
Sbjct: 181 GSQAGLSWLAKPVTGQSSIGLASRHGSIINQSMPLMDPLVTLFGSIHEKLPETGAGGSMR 360
Query: 317 STLFPHFGSMFSVGGNQPRNEDWDEESLAREGDDYISDAAAGDSDDNLQSPLISRQTTSM 376
STLFP+FGSMFS +NE WDEESL REG+DY+SDAA GDSDDNL SPLISRQTTS+
Sbjct: 361 STLFPNFGSMFSTAEPHAKNEQWDEESLQREGEDYMSDAAGGDSDDNLHSPLISRQTTSL 540
Query: 377 DKDM--PLPAQGS-LSNMRQGSLLQGNAGEPVGSTGIGGGWQLAWKWSEQEGPGGKKEG 432
+KD+ P P+ GS L +MR+ S L +GE GSTGIGGGWQLAWKW++ +G GK++G
Sbjct: 541 EKDLPPPPPSHGSILGSMRRHSSLMQGSGEQGGSTGIGGGWQLAWKWTD-KGEDGKQQG 714
>TC203480 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, partial (38%)
Length = 1040
Score = 207 bits (526), Expect = 1e-53
Identities = 107/146 (73%), Positives = 123/146 (83%)
Frame = +2
Query: 447 SIRASVVSLPGGDVPTDGDVVQQAAALVSQPALYNKELMHQQPVGPAMIHPSETAAKGPS 506
S R S+VS+PG +G+ VQ AAALVSQPALY+KEL+ PVGPAM+HPSETA+KGPS
Sbjct: 11 SHRGSIVSIPG-----EGEFVQ-AAALVSQPALYSKELIDGHPVGPAMVHPSETASKGPS 172
Query: 507 WNDLFEPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSF 566
W L EPGVKHAL VGVG+QILQQFSGINGVLYYTPQILE+AGV LLS++G+ S S+SF
Sbjct: 173 WKALLEPGVKHALVVGVGIQILQQFSGINGVLYYTPQILEEAGVEVLLSDIGIGSESASF 352
Query: 567 LISAVTTLLMLPCIAVAMRLMDISGR 592
LISA TT LMLPCI VAM+LMD+SGR
Sbjct: 353 LISAFTTFLMLPCIGVAMKLMDVSGR 430
>TC229015 similar to UP|Q8GT52 (Q8GT52) Hexose transporter, partial (27%)
Length = 618
Score = 119 bits (297), Expect(3) = 4e-46
Identities = 60/63 (95%), Positives = 62/63 (98%)
Frame = +1
Query: 530 QFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLISAVTTLLMLPCIAVAMRLMDI 589
QFSGINGVLYYTPQILEQAGVGYLLS+LGL STSSSFLISAVTTLLMLPCIA+AMRLMDI
Sbjct: 106 QFSGINGVLYYTPQILEQAGVGYLLSSLGLGSTSSSFLISAVTTLLMLPCIAIAMRLMDI 285
Query: 590 SGR 592
SGR
Sbjct: 286 SGR 294
Score = 64.7 bits (156), Expect(3) = 4e-46
Identities = 29/37 (78%), Positives = 33/37 (88%)
Frame = +2
Query: 498 SETAAKGPSWNDLFEPGVKHALFVGVGLQILQQFSGI 534
+ET AKGPSW+DLFEPGVKHAL VGVG+QILQQ S +
Sbjct: 5 AETIAKGPSWSDLFEPGVKHALIVGVGMQILQQCSSL 115
Score = 40.8 bits (94), Expect(3) = 4e-46
Identities = 16/24 (66%), Positives = 18/24 (74%)
Frame = +3
Query: 593 SISFHISPREPCGFGRHCKCVYLN 616
S S HISP + CGFG HCKC+ LN
Sbjct: 330 SSSSHISPGKSCGFGNHCKCINLN 401
>BF070135
Length = 422
Score = 137 bits (346), Expect = 1e-32
Identities = 75/108 (69%), Positives = 86/108 (79%), Gaps = 3/108 (2%)
Frame = +2
Query: 428 GKKEGGFKRIYL-HQEGGPGSIRASVVSLPGGDVPT--DGDVVQQAAALVSQPALYNKEL 484
G K+ F R Y+ ++ GS R S++SLPG D PT DG++VQ AAALVSQ ALYNKEL
Sbjct: 101 GLKQKVFSREYICTKKVVLGSRRGSIISLPGCDAPTLTDGEIVQ-AAALVSQSALYNKEL 277
Query: 485 MHQQPVGPAMIHPSETAAKGPSWNDLFEPGVKHALFVGVGLQILQQFS 532
MHQQPVGPAMIHPS+TAAKGPSW+DL EPGVKHAL V G+QILQQFS
Sbjct: 278 MHQQPVGPAMIHPSQTAAKGPSWSDLLEPGVKHALIVXGGIQILQQFS 421
Score = 87.0 bits (214), Expect = 2e-17
Identities = 46/65 (70%), Positives = 48/65 (73%)
Frame = +1
Query: 381 PLPAQGSLSNMRQGSLLQGNAGEPVGSTGIGGGWQLAWKWSEQEGPGGKKEGGFKRIYLH 440
P PAQGS MRQGSLLQG EP G++GIGGGWQLAWKWSE EG FKRIYLH
Sbjct: 1 PAPAQGS---MRQGSLLQG---EPAGNSGIGGGWQLAWKWSETEGV-------FKRIYLH 141
Query: 441 QEGGP 445
QEGGP
Sbjct: 142 QEGGP 156
>TC205503 UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, complete
Length = 1928
Score = 128 bits (321), Expect = 9e-30
Identities = 74/215 (34%), Positives = 126/215 (58%), Gaps = 3/215 (1%)
Frame = +1
Query: 4 AVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPT--VEGLIVAMSLIGATVVTT 61
A A+ A++ ++L G+D ++G+ +YIKR+ ++ E + G+I SLIG+ +
Sbjct: 256 AFACAMLASMTSILLGYDIGVMSGAAIYIKRDLKVSDEQIEILLGIINLYSLIGSCL--- 426
Query: 62 CSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYI 121
+G SD GRR + + ++ + S +M + P+ L+ R + G+GIG A+ + P+Y
Sbjct: 427 -AGRTSDWIGRRYTIGLGGAIFLVGSTLMGFYPHYSFLMCGRFVAGIGIGYALMIAPVYT 603
Query: 122 SEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMS-LTKAPSWRLMLGVLSIPSLIYFAL 180
+E++P RG L + P+ + G+ Y +G S LT WR+MLGV +IPS++
Sbjct: 604 AEVSPASSRGFLTSFPEVFINGGILLGYISNYGFSKLTLKVGWRMMLGVGAIPSVV-LTE 780
Query: 181 TLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVA 215
+L +PESPRWLV +GR+ EA+KVL + ++ A
Sbjct: 781 GVLAMPESPRWLVMRGRLGEARKVLNKTSDSKEEA 885
Score = 48.5 bits (114), Expect = 9e-06
Identities = 29/116 (25%), Positives = 58/116 (50%), Gaps = 24/116 (20%)
Frame = +1
Query: 507 WNDLF---EPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGV----GYLLSNLGL 559
W +LF P ++H + +G+ QQ SG++ V+ Y+P+I E+AG+ LL+ + +
Sbjct: 985 WKELFLYPTPAIRHIVIAALGIHFFQQASGVDAVVLYSPRIFEKAGITNDTHKLLATVAV 1164
Query: 560 SSTSSSFLISAVTTL-----------------LMLPCIAVAMRLMDISGRSISFHI 598
+ F+++A TL L L +A+++ ++D S R + + +
Sbjct: 1165 GFVKTVFILAATFTLDRVGRRPLLLSSVGGMVLSLLTLAISLTVIDHSERKLMWAV 1332
>TC207288 UP|Q7XA51 (Q7XA51) Monosaccharide transporter, complete
Length = 1813
Score = 93.6 bits (231), Expect = 2e-19
Identities = 58/179 (32%), Positives = 99/179 (54%), Gaps = 2/179 (1%)
Frame = +1
Query: 47 LIVAMSLIGATVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLD 106
L + A V+T + L+ GR+ +I+ +L + +++ + N+ +L+ R+L
Sbjct: 286 LFTSSLYFSALVMTFFASFLTRKKGRKASIIVGALSFLAGAILNAAAKNIAMLIIGRVLL 465
Query: 107 GLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLM 166
G GIG VPLY+SE+AP + RG++N L QF AG+ + + + WR+
Sbjct: 466 GGGIGFGNQAVPLYLSEMAPAKNRGAVNQLFQFTTCAGILIANLVNYFTEKIHPYGWRIS 645
Query: 167 LGVLSIPSLIYFALTL--LLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLVE 223
LG+ +P+ FA+ + + E+P LV +GR+ +AK+VLQR+RG ++V E L E
Sbjct: 646 LGLAGLPA---FAMLVGGICCAETPNSLVEQGRLDKAKQVLQRIRGTENVEAEFEDLKE 813
>TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein, partial
(93%)
Length = 1695
Score = 91.3 bits (225), Expect = 1e-18
Identities = 53/164 (32%), Positives = 89/164 (53%)
Frame = +2
Query: 47 LIVAMSLIGATVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLD 106
L ++S +GA V SG +++ GR+ L+I+S+ + L + ++ + L RLL+
Sbjct: 296 LFGSLSNVGAMVGAIASGQIAEYIGRKGSLMIASIPNIIGWLAISFAKDSSFLYMGRLLE 475
Query: 107 GLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLM 166
G G+G+ VP+YI+EI+PP +RG L ++ Q + + G+ +Y + + WR++
Sbjct: 476 GFGVGIISYTVPVYIAEISPPNLRGGLVSVNQLSVTIGIMLAYLLGIFV------EWRIL 637
Query: 167 LGVLSIPSLIYFALTLLLLPESPRWLVSKGRMLEAKKVLQRLRG 210
+ +P I L +PESPRWL G E + LQ LRG
Sbjct: 638 AIIGILPCTILIP-ALFFIPESPRWLAKMGMTEEFETSLQVLRG 766
Score = 50.1 bits (118), Expect = 3e-06
Identities = 31/81 (38%), Positives = 45/81 (55%)
Frame = +2
Query: 519 LFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLISAVTTLLMLP 578
L +G+GL ILQQ SGINGVL+Y+ I AG+ SS +++F + AV L
Sbjct: 881 LMIGIGLLILQQLSGINGVLFYSSTIFRNAGIS--------SSDAATFGVGAVQVL---- 1024
Query: 579 CIAVAMRLMDISGRSISFHIS 599
++ + L D SGR + +S
Sbjct: 1025ATSLTLWLADKSGRRLLLIVS 1087
>TC222070 weakly similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, partial
(17%)
Length = 376
Score = 86.7 bits (213), Expect = 3e-17
Identities = 45/64 (70%), Positives = 52/64 (80%), Gaps = 1/64 (1%)
Frame = +3
Query: 538 LYYTPQILEQAGVGYLLSNLGLSSTSSSFLISAVTTLLMLPCIAVAMRLMDISG-RSISF 596
LYY PQILE AGVG LLSNLGLSS S+SFL++ +TT MLPCIA+A+RLMDISG RSI
Sbjct: 6 LYYAPQILEXAGVGALLSNLGLSSASASFLVNIITTFCMLPCIALAVRLMDISGRRSIML 185
Query: 597 HISP 600
+ P
Sbjct: 186 YTVP 197
>BF009879 weakly similar to GP|26986186|em hexose transporter {Hordeum
vulgare subsp. vulgare}, partial (14%)
Length = 595
Score = 80.1 bits (196), Expect(2) = 2e-16
Identities = 41/57 (71%), Positives = 48/57 (83%)
Frame = +2
Query: 536 GVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLISAVTTLLMLPCIAVAMRLMDISGR 592
GVLYYTPQILEQA V YLLS L L+STSSSFLIS TTLL+LP IA+ ++L++IS R
Sbjct: 2 GVLYYTPQILEQASVNYLLSTLSLNSTSSSFLISTRTTLLILPYIAITIKLINISSR 172
Score = 24.3 bits (51), Expect(2) = 2e-16
Identities = 11/24 (45%), Positives = 14/24 (57%)
Frame = +1
Query: 593 SISFHISPREPCGFGRHCKCVYLN 616
S S HI+PR+ C H K + LN
Sbjct: 208 SSSSHITPRKSCELTIHYKYINLN 279
>TC233090 similar to UP|Q9ZVN7 (Q9ZVN7) T7A14.10 protein, partial (22%)
Length = 594
Score = 83.2 bits (204), Expect = 3e-16
Identities = 48/155 (30%), Positives = 83/155 (52%)
Frame = +3
Query: 9 VAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVTTCSGALSD 68
+ A++ N + G+ + G I+ I RE + +EGL+V++ + GA + + S +L D
Sbjct: 15 LVASMSNFIFGYHIGVMNGPIVSIARELGFEGNSFIEGLVVSIFIAGAFIGSISSASLLD 194
Query: 69 LFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIAPPE 128
G R I+S+ L +++ + ++ ++ R L GLGIG+ LVP+YISE+AP +
Sbjct: 195 RLGSRLTFQINSIPLILGAIISAQAHSLNEIIGGRFLVGLGIGVNTVLVPIYISEVAPTK 374
Query: 129 IRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSW 163
RG+L +L Q G+ S + G+ P W
Sbjct: 375 YRGALGSLCQIGTCLGIITS--LFLGIPSENDPHW 473
>BE661114
Length = 634
Score = 82.8 bits (203), Expect = 4e-16
Identities = 43/118 (36%), Positives = 74/118 (62%), Gaps = 1/118 (0%)
Frame = +3
Query: 7 VAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPT-VEGLIVAMSLIGATVVTTCSGA 65
+A +A IG LL G+D I+G++LYIK EF+ T ++ IV+ ++ GA + + G
Sbjct: 261 LAFSAGIGGLLFGYDTGVISGALLYIKDEFKAVDRKTWLQEAIVSTAIAGAIIGASVGGW 440
Query: 66 LSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISE 123
++D FGR+ ++I+ L+F+ S++M + + IL+ R+ G+G+ +A PLYISE
Sbjct: 441 INDRFGRKKGIVIADTLFFIGSVIMAAASSPAILIVGRVFVGIGVXMASMASPLYISE 614
>TC233218 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, partial (5%)
Length = 622
Score = 75.1 bits (183), Expect = 9e-14
Identities = 37/38 (97%), Positives = 38/38 (99%)
Frame = +3
Query: 1 MSGAVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQL 38
MSGAV+VAVAAAIGNLLQGWDNATIAGSILYIKREFQL
Sbjct: 507 MSGAVLVAVAAAIGNLLQGWDNATIAGSILYIKREFQL 620
>TC206001 similar to UP|Q8VZI5 (Q8VZI5) AT5g18840/F17K4_90, partial (66%)
Length = 951
Score = 71.6 bits (174), Expect = 9e-13
Identities = 49/137 (35%), Positives = 68/137 (48%), Gaps = 1/137 (0%)
Frame = +3
Query: 88 LVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFF 147
L +F+S Y L R G GIG+ +VP+YI+EIAP +RG L T Q G
Sbjct: 6 LAVFFSKGSYSLDMGRFFTGYGIGVISYVVPVYIAEIAPKNLRGGLATTNQLLIVTGGSV 185
Query: 148 SYCMVFGMSLTKAPSWRLMLGVLSIPSLIYFALTLLLLPESPRWLVSKGRMLEAKKVLQR 207
S+ L +WR L + + I + L +PESPRWL GR E + L R
Sbjct: 186 SFL------LGSVINWR-ELALAGLVPCICLLVGLCFIPESPRWLAKVGREKEFQLALSR 344
Query: 208 LRGCQ-DVAGEMALLVE 223
LRG D++ E A +++
Sbjct: 345 LRGKHADISDEAAEILD 395
Score = 44.7 bits (104), Expect = 1e-04
Identities = 30/84 (35%), Positives = 42/84 (49%)
Frame = +3
Query: 509 DLFEPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLI 568
DLF+ H++ +GVGL QQ GING+ +YT +I A GLSS + +
Sbjct: 438 DLFQSKYVHSVVIGVGLMACQQSVGINGIGFYTAEIFVAA---------GLSSGKAGTIA 590
Query: 569 SAVTTLLMLPCIAVAMRLMDISGR 592
A + +P + LMD SGR
Sbjct: 591 YA---CIQIPFTLLGAILMDKSGR 653
>BE330114 similar to PIR|T45780|T457 sugar transporter-like protein -
Arabidopsis thaliana, partial (3%)
Length = 226
Score = 71.2 bits (173), Expect = 1e-12
Identities = 37/73 (50%), Positives = 49/73 (66%), Gaps = 3/73 (4%)
Frame = +2
Query: 358 GDSDDNLQSPLISRQTTSMDKDMPLPA--QGS-LSNMRQGSLLQGNAGEPVGSTGIGGGW 414
GD DD + +IS QTTS++KD+P PA GS L +MR+ S L +GE G+T IGGGW
Sbjct: 2 GDCDDY*HNTIISLQTTSLEKDLPAPAASHGSILGSMRRDSGLMQGSGEQCGNTRIGGGW 181
Query: 415 QLAWKWSEQEGPG 427
LAWKW+++ G
Sbjct: 182 HLAWKWTDKGEDG 220
>TC229014 similar to UP|Q8GT52 (Q8GT52) Hexose transporter, partial (23%)
Length = 714
Score = 47.8 bits (112), Expect(2) = 3e-12
Identities = 25/28 (89%), Positives = 26/28 (92%)
Frame = +2
Query: 565 SFLISAVTTLLMLPCIAVAMRLMDISGR 592
SFLISAVTTLLML IA+AMRLMDISGR
Sbjct: 2 SFLISAVTTLLMLXXIAIAMRLMDISGR 85
Score = 42.4 bits (98), Expect(2) = 3e-12
Identities = 17/24 (70%), Positives = 18/24 (74%)
Frame = +1
Query: 593 SISFHISPREPCGFGRHCKCVYLN 616
S S HISP E CGFG HCKC+ LN
Sbjct: 121 SSSSHISPGESCGFGNHCKCINLN 192
>TC216287 similar to UP|Q8GTR0 (Q8GTR0) Sugar transporter, partial (37%)
Length = 696
Score = 65.1 bits (157), Expect = 9e-11
Identities = 39/128 (30%), Positives = 66/128 (51%), Gaps = 1/128 (0%)
Frame = +3
Query: 95 NVYILLFARLLDGLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFG 154
N++ +L RL G G+GL + LY++E++P +RG+ Q A G+ + + G
Sbjct: 39 NLFGMLVGRLFVGTGLGLGPPVASLYVTEVSPAFVRGTFGAFIQIATCLGLMGA--LFIG 212
Query: 155 MSLTKAPS-WRLMLGVLSIPSLIYFALTLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQD 213
+ + + WR+ V +IP+ I A ++ ESP WL +GR EA+ +RL G +
Sbjct: 213 IPVKEISGWWRVCFWVSTIPAAI-LATAMVFCAESPHWLYKQGRTAEAEAEFERLLGVSE 389
Query: 214 VAGEMALL 221
M+ L
Sbjct: 390 AKFAMSEL 413
Score = 28.5 bits (62), Expect = 9.2
Identities = 12/26 (46%), Positives = 17/26 (65%)
Frame = +3
Query: 519 LFVGVGLQILQQFSGINGVLYYTPQI 544
+F+G L LQQ SGIN V Y++ +
Sbjct: 618 VFIGSTLFALQQLSGINAVFYFSSTV 695
>TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 2338
Score = 59.7 bits (143), Expect = 4e-09
Identities = 32/72 (44%), Positives = 47/72 (64%), Gaps = 1/72 (1%)
Frame = +3
Query: 145 MFFSYCMVFGMS-LTKAPSWRLMLGVLSIPSLIYFALTLLLLPESPRWLVSKGRMLEAKK 203
+ Y +G S L WRLMLGV +IPS++ + +L +PESPRWLV+KGR+ EAK+
Sbjct: 3 ILIGYISNYGFSKLALRLGWRLMLGVGAIPSIL-IGVAVLAMPESPRWLVAKGRLGEAKR 179
Query: 204 VLQRLRGCQDVA 215
VL ++ ++ A
Sbjct: 180 VLYKISESEEEA 215
Score = 42.7 bits (99), Expect = 5e-04
Identities = 25/91 (27%), Positives = 42/91 (45%), Gaps = 3/91 (3%)
Frame = +3
Query: 507 WNDLF---EPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTS 563
W +LF P V+H +G+ Q +GI+ V+ Y+P+I E+AG+ S +
Sbjct: 315 WRELFLHPTPAVRHIFIASLGIHFFAQATGIDAVVLYSPRIFEKAGI---------KSDN 467
Query: 564 SSFLISAVTTLLMLPCIAVAMRLMDISGRSI 594
L + + I VA +D +GR +
Sbjct: 468 YRLLATVAVGFVKTVSILVATFFLDRAGRRV 560
>BE555798 homologue to GP|1353516|gb|A sugar transporter {Medicago
truncatula}, partial (18%)
Length = 312
Score = 57.0 bits (136), Expect = 2e-08
Identities = 29/76 (38%), Positives = 50/76 (65%)
Frame = +2
Query: 70 FGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIAPPEI 129
FGR+ ++ LL+ + +L+ ++ +V++L+ R+L G GIG A VPLY+SE+AP +
Sbjct: 11 FGRKLSMLF*GLLFLVGALINGFAQHVWMLIVGRILLGFGIGFANQSVPLYLSEMAPYKY 190
Query: 130 RGSLNTLPQFAGSAGM 145
RG+LN Q + + G+
Sbjct: 191 RGALNIGFQLSITVGI 238
>CA952974 similar to GP|17381265|gb| AT5g18840/F17K4_90 {Arabidopsis
thaliana}, partial (19%)
Length = 372
Score = 55.8 bits (133), Expect = 5e-08
Identities = 33/85 (38%), Positives = 45/85 (52%)
Frame = +3
Query: 54 IGATVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLA 113
IGA + SG ++D GR+ + IS+ LV+F+S Y L R G GIG+
Sbjct: 39 IGAMLGAISSGRITDFIGRKGAMRISAGFCITGWLVVFFSKGSYSLDLGRFFTGYGIGVI 218
Query: 114 VTLVPLYISEIAPPEIRGSLNTLPQ 138
+VP+YI EIAP +R L T Q
Sbjct: 219 SYVVPVYIVEIAPKNLREELATTNQ 293
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,633,085
Number of Sequences: 63676
Number of extensions: 398572
Number of successful extensions: 2446
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 2390
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2420
length of query: 616
length of database: 12,639,632
effective HSP length: 103
effective length of query: 513
effective length of database: 6,081,004
effective search space: 3119555052
effective search space used: 3119555052
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC121239.3


