

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121238.7 - phase: 0
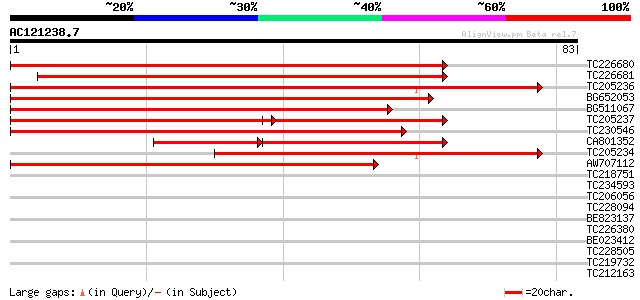
(83 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226680 weakly similar to UP|SCS2_YEAST (P40075) SCS2 protein, ... 99 5e-22
TC226681 similar to GB|AAQ63968.1|34329673|AY364005 VAP27-1 {Ara... 94 1e-20
TC205236 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (82%) 77 2e-15
BG652053 similar to GP|20160761|dbj P0504E02.2 {Oryza sativa (ja... 70 2e-13
BG511067 weakly similar to PIR|B86220|B86 protein F22O13.31 [imp... 68 7e-13
TC205237 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (58%) 56 2e-12
TC230546 similar to GB|AAM62506.1|21553413|AY085274 VAMP (vesicl... 62 5e-11
CA801352 similar to PIR|JC7234|JC72 27k vesicle-associated membr... 36 6e-06
TC205234 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (56%) 44 2e-05
AW707112 39 4e-04
TC218751 similar to UP|Q6W6U5 (Q6W6U5) Olfactory receptor family... 32 0.074
TC234593 similar to UP|Q9LPZ8 (Q9LPZ8) T23J18.3, partial (12%) 29 0.48
TC206056 similar to UP|Q948Y6 (Q948Y6) VMP4 protein, partial (3%) 28 0.82
TC228094 similar to UP|Q8RUF8 (Q8RUF8) AT5g12040/F14F18_210, par... 27 1.8
BE823137 similar to SP|Q9ZW96|LB40_ LOB domain protein 40. [Mous... 27 1.8
TC226380 similar to UP|Q6TUE7 (Q6TUE7) LRRGT00097, partial (3%) 27 2.4
BE023412 27 2.4
TC228505 similar to UP|Q7XA73 (Q7XA73) At4g14710/dl3395c, partia... 27 2.4
TC219732 similar to UP|PEAM_SPIOL (Q9M571) Phosphoethanolamine N... 26 3.1
TC212163 similar to UP|Q9LT26 (Q9LT26) Arabidopsis thaliana geno... 26 3.1
>TC226680 weakly similar to UP|SCS2_YEAST (P40075) SCS2 protein, partial
(16%)
Length = 1112
Score = 98.6 bits (244), Expect = 5e-22
Identities = 46/64 (71%), Positives = 54/64 (83%)
Frame = +3
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPP 60
MQA +EAPPDMQCKDKFLLQS+VA PGATTKDIT +MF K+SG++V+E K R V VAPP
Sbjct: 336 MQAQKEAPPDMQCKDKFLLQSVVASPGATTKDITPEMFNKESGHDVEECKLRVVYVAPPQ 515
Query: 61 SPNP 64
P+P
Sbjct: 516 PPSP 527
>TC226681 similar to GB|AAQ63968.1|34329673|AY364005 VAP27-1 {Arabidopsis
thaliana;} , partial (39%)
Length = 829
Score = 94.0 bits (232), Expect = 1e-20
Identities = 43/60 (71%), Positives = 51/60 (84%)
Frame = +3
Query: 5 EEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPPSPNP 64
+EAPPDMQCKDKFLLQS+VA PGATTKDIT +MF K+SG++V+E K R V VAPP P+P
Sbjct: 54 KEAPPDMQCKDKFLLQSVVASPGATTKDITPEMFNKESGHDVEECKLRVVYVAPPQPPSP 233
>TC205236 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (82%)
Length = 765
Score = 77.0 bits (188), Expect = 2e-15
Identities = 42/80 (52%), Positives = 52/80 (64%), Gaps = 2/80 (2%)
Frame = +2
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAP-- 58
MQA +EAP DMQCKDKFLLQS+ G + KDIT+DMF K++G+ V+E K R + V+P
Sbjct: 344 MQAQKEAPADMQCKDKFLLQSVKTVDGTSPKDITADMFNKEAGHVVEECKLRVLYVSPP* 523
Query: 59 PPSPNPPLSVHASESLDQVS 78
PPS P S S VS
Sbjct: 524 PPSSVPEGSEEGSSPRGSVS 583
>BG652053 similar to GP|20160761|dbj P0504E02.2 {Oryza sativa (japonica
cultivar-group)}, partial (12%)
Length = 412
Score = 69.7 bits (169), Expect = 2e-13
Identities = 34/62 (54%), Positives = 40/62 (63%)
Frame = +3
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPP 60
MQA APPD+ CKDKFL+QS V G T +I+SD+F KDSG V E K R V + P
Sbjct: 84 MQAQRTAPPDLHCKDKFLVQSAVVPKGTTEDEISSDLFVKDSGRLVDEKKLRVVLINSPS 263
Query: 61 SP 62
SP
Sbjct: 264 SP 269
>BG511067 weakly similar to PIR|B86220|B86 protein F22O13.31 [imported] -
Arabidopsis thaliana, partial (27%)
Length = 408
Score = 68.2 bits (165), Expect = 7e-13
Identities = 34/56 (60%), Positives = 38/56 (67%)
Frame = +2
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCV 56
MQA APPDM CKDKFL+QS V G T DITSDMF KDSG ++E K R V +
Sbjct: 239 MQAQRMAPPDMLCKDKFLIQSTVVPFGTTEDDITSDMFSKDSGKYIEEKKLRVVLI 406
>TC205237 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (58%)
Length = 696
Score = 55.8 bits (133), Expect(2) = 2e-12
Identities = 26/39 (66%), Positives = 30/39 (76%)
Frame = +3
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFY 39
MQA +EAP DMQCKDKFLLQS+ G + KDIT+DM Y
Sbjct: 375 MQAQKEAPADMQCKDKFLLQSVKTVDGTSPKDITADMVY 491
Score = 30.8 bits (68), Expect(2) = 2e-12
Identities = 12/27 (44%), Positives = 19/27 (69%)
Frame = +1
Query: 38 FYKDSGYEVKESKFRAVCVAPPPSPNP 64
F K++G+ V+E K R + V+PP P+P
Sbjct: 583 FNKEAGHVVEECKLRVLYVSPPQPPSP 663
>TC230546 similar to GB|AAM62506.1|21553413|AY085274 VAMP (vesicle-associated
membrane protein)-associated protein-like {Arabidopsis
thaliana;} , partial (60%)
Length = 1019
Score = 62.0 bits (149), Expect = 5e-11
Identities = 27/58 (46%), Positives = 36/58 (61%)
Frame = +3
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAP 58
+QA +E PPDMQCKDKFLLQS + P D+ D F KD +++ K R V ++P
Sbjct: 423 LQAQQEYPPDMQCKDKFLLQSTIVNPNTDVDDLPPDTFNKDGEKSIEDMKLRVVYISP 596
>CA801352 similar to PIR|JC7234|JC72 27k vesicle-associated membrane
protein-associated protein - curled-leaved tobacco,
partial (25%)
Length = 400
Score = 35.8 bits (81), Expect(2) = 6e-06
Identities = 15/27 (55%), Positives = 20/27 (73%)
Frame = +3
Query: 38 FYKDSGYEVKESKFRAVCVAPPPSPNP 64
F K+SG++V+E K R V VAPP P+P
Sbjct: 144 FNKESGHDVEECKLRVVYVAPPQPPSP 224
Score = 28.9 bits (63), Expect(2) = 6e-06
Identities = 12/16 (75%), Positives = 14/16 (87%)
Frame = +2
Query: 22 IVARPGATTKDITSDM 37
+VA PGATTKDIT +M
Sbjct: 2 VVASPGATTKDITPEM 49
>TC205234 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (56%)
Length = 948
Score = 43.5 bits (101), Expect = 2e-05
Identities = 24/50 (48%), Positives = 31/50 (62%), Gaps = 2/50 (4%)
Frame = +1
Query: 31 KDITSDMFYKDSGYEVKESKFRAVCVAP--PPSPNPPLSVHASESLDQVS 78
KDIT+DMF K++G+ V+E K R + V+P PPSP P S S VS
Sbjct: 73 KDITADMFNKEAGHVVEECKLRVLYVSPPQPPSPVPEGSEEGSSPRGSVS 222
>AW707112
Length = 433
Score = 39.3 bits (90), Expect = 4e-04
Identities = 20/54 (37%), Positives = 35/54 (64%)
Frame = +2
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAV 54
+Q +E+P ++Q ++KFLL+SI K+IT+DMF K + + V++ K + V
Sbjct: 197 IQTQKESPTNIQYENKFLLESIKTIDNTXPKNITTDMFNKRTEHIVEKCK*KVV 358
>TC218751 similar to UP|Q6W6U5 (Q6W6U5) Olfactory receptor family 8 subfamily
S (Fragment), partial (7%)
Length = 988
Score = 31.6 bits (70), Expect = 0.074
Identities = 13/25 (52%), Positives = 17/25 (68%)
Frame = +2
Query: 38 FYKDSGYEVKESKFRAVCVAPPPSP 62
F KDSG ++E K R V ++PP SP
Sbjct: 185 FAKDSGKFIEEKKLRVVLISPPSSP 259
>TC234593 similar to UP|Q9LPZ8 (Q9LPZ8) T23J18.3, partial (12%)
Length = 532
Score = 28.9 bits (63), Expect = 0.48
Identities = 11/22 (50%), Positives = 14/22 (63%)
Frame = +2
Query: 52 RAVCVAPPPSPNPPLSVHASES 73
RA+ PPP P PP HA++S
Sbjct: 131 RAITTPPPPPPPPPPPAHAAKS 196
>TC206056 similar to UP|Q948Y6 (Q948Y6) VMP4 protein, partial (3%)
Length = 781
Score = 28.1 bits (61), Expect = 0.82
Identities = 10/13 (76%), Positives = 12/13 (91%)
Frame = +3
Query: 57 APPPSPNPPLSVH 69
+PPPSP+PPLS H
Sbjct: 87 SPPPSPSPPLSHH 125
>TC228094 similar to UP|Q8RUF8 (Q8RUF8) AT5g12040/F14F18_210, partial (82%)
Length = 1538
Score = 26.9 bits (58), Expect = 1.8
Identities = 9/15 (60%), Positives = 12/15 (80%)
Frame = +3
Query: 53 AVCVAPPPSPNPPLS 67
++ + PPP PNPPLS
Sbjct: 135 SLSIPPPPPPNPPLS 179
>BE823137 similar to SP|Q9ZW96|LB40_ LOB domain protein 40. [Mouse-ear cress]
{Arabidopsis thaliana}, partial (10%)
Length = 572
Score = 26.9 bits (58), Expect = 1.8
Identities = 13/28 (46%), Positives = 17/28 (60%), Gaps = 4/28 (14%)
Frame = +3
Query: 50 KFRAVCVAPPPSPNPPLS----VHASES 73
+FR++C PPPSP P S H S+S
Sbjct: 333 RFRSLCFPPPPSPWLPTSGSAPTHDSQS 416
>TC226380 similar to UP|Q6TUE7 (Q6TUE7) LRRGT00097, partial (3%)
Length = 715
Score = 26.6 bits (57), Expect = 2.4
Identities = 20/78 (25%), Positives = 29/78 (36%), Gaps = 7/78 (8%)
Frame = +3
Query: 10 DMQCKDKFLLQSIVARPGATTKDITS----DMFYKDSGYEV---KESKFRAVCVAPPPSP 62
D + Q+ GAT D+ ++ D Y KESK A PP
Sbjct: 159 DRNAVSSVITQTETETDGATIVDVAVAVNVNVNVNDENYRTPTSKESKIPATMTCPPAPR 338
Query: 63 NPPLSVHASESLDQVSFY 80
P L+ + LD+ F+
Sbjct: 339 KPKLASCKRKLLDEFQFF 392
>BE023412
Length = 294
Score = 26.6 bits (57), Expect = 2.4
Identities = 9/13 (69%), Positives = 11/13 (84%)
Frame = -1
Query: 55 CVAPPPSPNPPLS 67
C+ PPPS NPPL+
Sbjct: 198 CLHPPPSQNPPLT 160
>TC228505 similar to UP|Q7XA73 (Q7XA73) At4g14710/dl3395c, partial (28%)
Length = 913
Score = 26.6 bits (57), Expect = 2.4
Identities = 11/23 (47%), Positives = 15/23 (64%)
Frame = +2
Query: 57 APPPSPNPPLSVHASESLDQVSF 79
APPP P+ P S S SL +++F
Sbjct: 167 APPPPPSDPSSTSPSTSLPRMTF 235
>TC219732 similar to UP|PEAM_SPIOL (Q9M571) Phosphoethanolamine
N-methyltransferase , partial (48%)
Length = 812
Score = 26.2 bits (56), Expect = 3.1
Identities = 13/31 (41%), Positives = 17/31 (53%)
Frame = -1
Query: 37 MFYKDSGYEVKESKFRAVCVAPPPSPNPPLS 67
MF + S SKF A+ ++PPP P P S
Sbjct: 188 MFMERSMPTTSTSKFSAM*MSPPPVPQPTSS 96
>TC212163 similar to UP|Q9LT26 (Q9LT26) Arabidopsis thaliana genomic DNA,
chromosome 3, P1 clone: MPN9, partial (30%)
Length = 739
Score = 26.2 bits (56), Expect = 3.1
Identities = 10/24 (41%), Positives = 16/24 (66%)
Frame = -3
Query: 53 AVCVAPPPSPNPPLSVHASESLDQ 76
AV +APPP P+P L H ++ ++
Sbjct: 284 AVVLAPPPPPSPQLVEHDADEEEE 213
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,261,371
Number of Sequences: 63676
Number of extensions: 60582
Number of successful extensions: 1499
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 1174
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1488
length of query: 83
length of database: 12,639,632
effective HSP length: 59
effective length of query: 24
effective length of database: 8,882,748
effective search space: 213185952
effective search space used: 213185952
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Medicago: description of AC121238.7


