

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121237.18 - phase: 0
(387 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
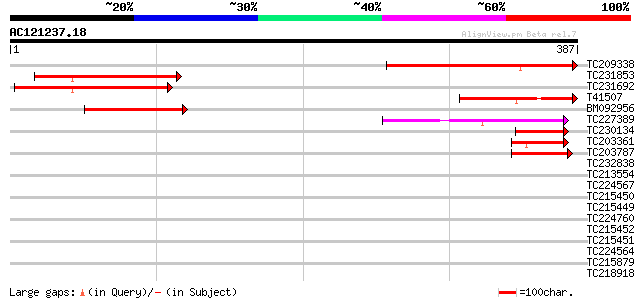
Score E
Sequences producing significant alignments: (bits) Value
TC209338 weakly similar to UP|Q8W586 (Q8W586) AT4g21800/F17L22_2... 211 4e-55
TC231853 similar to UP|Q8W586 (Q8W586) AT4g21800/F17L22_260, par... 171 5e-43
TC231692 similar to UP|Q8W586 (Q8W586) AT4g21800/F17L22_260, par... 167 7e-42
T41507 similar to SP|Q9NVW2|RN12_ RING finger protein 12 (LIM do... 101 5e-22
BM092956 59 4e-09
TC227389 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacety... 44 9e-05
TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Pro... 44 1e-04
TC203361 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%) 43 3e-04
TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 42 5e-04
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 40 0.002
TC213554 weakly similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protei... 40 0.002
TC224567 similar to PIR|T51159|T51159 HMG protein [imported] - A... 40 0.002
TC215450 similar to UP|P93488 (P93488) NAP1Ps, partial (30%) 39 0.004
TC215449 similar to UP|P93488 (P93488) NAP1Ps, partial (97%) 39 0.004
TC224760 similar to UP|O04692 (O04692) DNA-binding protein, part... 39 0.004
TC215452 similar to UP|P93488 (P93488) NAP1Ps, partial (29%) 39 0.005
TC215451 weakly similar to UP|Q94K07 (Q94K07) Nucleosome assembl... 39 0.005
TC224564 similar to PIR|T51159|T51159 HMG protein [imported] - A... 39 0.005
TC215879 similar to UP|IF5_PHAVU (P48724) Eukaryotic translation... 38 0.009
TC218918 UP|Q39892 (Q39892) Nucleosome assembly protein 1, complete 37 0.012
>TC209338 weakly similar to UP|Q8W586 (Q8W586) AT4g21800/F17L22_260, partial
(28%)
Length = 544
Score = 211 bits (537), Expect = 4e-55
Identities = 106/135 (78%), Positives = 119/135 (87%), Gaps = 5/135 (3%)
Frame = +3
Query: 258 LALDEFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRR 317
L LDEFY+NL+S GVSAV+G G+E FF AVEASAEEYMETYKADLDKRREEKQ LEE+RR
Sbjct: 6 LVLDEFYNNLKSVGVSAVSGVGMEAFFNAVEASAEEYMETYKADLDKRREEKQRLEEDRR 185
Query: 318 KENMDKLRREMEKSGGETVVLSTGLKNKEE-----DEDEEDEEMDDDDNVDFGTYTEDED 372
KE+MDKLRR+MEKSGGETVVLSTGLK+KE DE++EDEEMDDDD+ D G YTE+ED
Sbjct: 186 KESMDKLRRDMEKSGGETVVLSTGLKDKEASKSMMDEEDEDEEMDDDDDDDLGIYTEEED 365
Query: 373 AIDEDEDEEVDKFSF 387
AIDEDEDEEVD+F F
Sbjct: 366 AIDEDEDEEVDRFGF 410
>TC231853 similar to UP|Q8W586 (Q8W586) AT4g21800/F17L22_260, partial (21%)
Length = 508
Score = 171 bits (433), Expect = 5e-43
Identities = 85/102 (83%), Positives = 94/102 (91%), Gaps = 2/102 (1%)
Frame = +3
Query: 18 KNKQKEELSESMKKLDIEGSSSGS--PNFKRKPVIIIVVGMAGSGKTTLMHRLVTHTHLS 75
K+K+KEEL+E+M KL IEGSSSGS N +RKPVII+VVGMAGSGKTTLMHRLV HTHL
Sbjct: 201 KDKEKEELTENMNKLHIEGSSSGSGSSNIRRKPVIILVVGMAGSGKTTLMHRLVCHTHLK 380
Query: 76 NIRGYVMNLDPAVMTLPYASNIDIRDTVKYKEVMKQFNLGPN 117
+IRGYVMNLDPAVMTLPYA+NID+RDTVKYKEVMKQFNLGPN
Sbjct: 381 DIRGYVMNLDPAVMTLPYAANIDVRDTVKYKEVMKQFNLGPN 506
>TC231692 similar to UP|Q8W586 (Q8W586) AT4g21800/F17L22_260, partial (19%)
Length = 735
Score = 167 bits (423), Expect = 7e-42
Identities = 84/110 (76%), Positives = 96/110 (86%), Gaps = 2/110 (1%)
Frame = +1
Query: 4 FFFFFFFFFLVLKEKNKQKEELSESMKKLDIEGSSSGS--PNFKRKPVIIIVVGMAGSGK 61
FF +F +LK K+K+KEEL+E+M KL IEGSSSGS +F+RKPVIIIVVGMAGSGK
Sbjct: 406 FFMLILWFCSILKAKDKEKEELTENMNKLHIEGSSSGSGSSSFRRKPVIIIVVGMAGSGK 585
Query: 62 TTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYKEVMKQ 111
TTLMHRLV HTHL +IRGYV+NLDPAVMTLPYA+NID+RDTVKYKEVMKQ
Sbjct: 586 TTLMHRLVCHTHLKDIRGYVVNLDPAVMTLPYAANIDVRDTVKYKEVMKQ 735
>T41507 similar to SP|Q9NVW2|RN12_ RING finger protein 12 (LIM domain
interacting RING finger protein), partial (5%)
Length = 411
Score = 101 bits (252), Expect = 5e-22
Identities = 55/88 (62%), Positives = 62/88 (69%), Gaps = 8/88 (9%)
Frame = -3
Query: 308 EKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKN--------KEEDEDEEDEEMDDD 359
E LEE R +E+MD LR +MEKSGGETVVLS G K EEDE+ EDEEMDDD
Sbjct: 409 ENPRLEEARGRESMDXLRXDMEKSGGETVVLSXGXKXXEASXXMMDEEDEEGEDEEMDDD 230
Query: 360 DNVDFGTYTEDEDAIDEDEDEEVDKFSF 387
D D G YTE+EDAIDE EDEE+D+F F
Sbjct: 229 D--DLGIYTEEEDAIDEGEDEEIDRFGF 152
>BM092956
Length = 421
Score = 58.9 bits (141), Expect = 4e-09
Identities = 28/70 (40%), Positives = 42/70 (60%)
Frame = +2
Query: 52 IVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYKEVMKQ 111
+V+G AGSGK+T L H + +V+NLDPA Y +DIR+ + +VM++
Sbjct: 197 LVIGPAGSGKSTYCSSLYEHCVAARRSIHVVNLDPAAENFDYPVAMDIRELISLDDVMEE 376
Query: 112 FNLGPNGGIL 121
LGPNGG++
Sbjct: 377 LGLGPNGGLV 406
>TC227389 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (63%)
Length = 1144
Score = 44.3 bits (103), Expect = 9e-05
Identities = 34/130 (26%), Positives = 63/130 (48%), Gaps = 3/130 (2%)
Frame = +3
Query: 255 SLSLALDEFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLE- 313
SL L L++ + S+ ++V G + ++ A + +E+ D D+ E+ L+
Sbjct: 261 SLELVLEKEFELSHSSKSASVHFCGYKAYYDADNSDEDEF-----TDSDEDDEDVPLINT 425
Query: 314 ENRRKENM--DKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDE 371
EN + E D E +K+ + + +K E +D E++ DD D+ DFG+ E+
Sbjct: 426 ENGKPETKAEDLKVPESKKAVAKASGSAKQVKVVEPKKDNEEDSDDDSDDDDFGSSDEEM 605
Query: 372 DAIDEDEDEE 381
+ D D D+E
Sbjct: 606 EDADSDSDDE 635
>TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23),
partial (8%)
Length = 581
Score = 43.9 bits (102), Expect = 1e-04
Identities = 19/36 (52%), Positives = 26/36 (71%)
Frame = +2
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
E D+D+ED++ DDDDN + +D+D DEDEDEE
Sbjct: 419 ESDDDDEDDDDDDDDNDEDDGDEDDDDEEDEDEDEE 526
Score = 35.8 bits (81), Expect = 0.034
Identities = 24/89 (26%), Positives = 45/89 (49%), Gaps = 6/89 (6%)
Frame = +2
Query: 302 LDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDED------EEDEE 355
LD+ + K EEN+ + + + + + GE G + E+DED E+ +
Sbjct: 221 LDQPCKGKHTSEENKDASDTEDDDDDDDVNDGED-----GDDDDEDDEDFSGDDGGEEAD 385
Query: 356 MDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
DDD + G ++D+D D+D+D++ D+
Sbjct: 386 SDDDPEANGGGESDDDDEDDDDDDDDNDE 472
Score = 32.3 bits (72), Expect = 0.37
Identities = 15/35 (42%), Positives = 23/35 (64%)
Frame = +2
Query: 350 DEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+ +D++ DDDD+ D DED DED+D+E D+
Sbjct: 419 ESDDDDEDDDDDDD----DNDEDDGDEDDDDEEDE 511
Score = 28.9 bits (63), Expect = 4.1
Identities = 17/44 (38%), Positives = 24/44 (53%), Gaps = 5/44 (11%)
Frame = +2
Query: 345 KEEDEDEEDE-----EMDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
+E D D++ E E DDDD D +D+D DED+ +E D
Sbjct: 374 EEADSDDDPEANGGGESDDDDEDD----DDDDDDNDEDDGDEDD 493
>TC203361 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%)
Length = 1090
Score = 42.7 bits (99), Expect = 3e-04
Identities = 22/47 (46%), Positives = 29/47 (60%), Gaps = 8/47 (17%)
Frame = -1
Query: 343 KNKEEDEDE--------EDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
+N EEDEDE ED++ DDDD D G EDED +++E+EE
Sbjct: 427 ENGEEDEDEDGDDQDDDEDDDDDDDDEEDEGGEEEDEDGAEDEENEE 287
Score = 33.1 bits (74), Expect = 0.22
Identities = 17/35 (48%), Positives = 21/35 (59%)
Frame = -1
Query: 347 EDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
E+ EEDE+ D DD D DED D+D+DEE
Sbjct: 430 EENGEEDEDEDGDDQDD------DEDDDDDDDDEE 344
Score = 32.7 bits (73), Expect = 0.29
Identities = 13/36 (36%), Positives = 24/36 (66%)
Frame = -1
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
E+D+D++D+E D+ + EDE+ +++EDEE
Sbjct: 376 EDDDDDDDDEEDEGGEEEDEDGAEDEENEEDEEDEE 269
Score = 30.8 bits (68), Expect = 1.1
Identities = 17/45 (37%), Positives = 22/45 (48%)
Frame = -1
Query: 333 GETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDED 377
GE S + E ED D+E DDDD+ D G +E+ ED
Sbjct: 640 GERKTQSQDKQRDAEGEDGNDDEGDDDDDGDGGFGEGEEELSSED 506
Score = 29.6 bits (65), Expect = 2.4
Identities = 10/36 (27%), Positives = 24/36 (65%)
Frame = -1
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
++D+D+++E+ ++ + G E+ + +EDE+EE
Sbjct: 370 DDDDDDDEEDEGGEEEDEDGAEDEENEEDEEDEEEE 263
>TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 1056
Score = 42.0 bits (97), Expect = 5e-04
Identities = 20/42 (47%), Positives = 26/42 (61%)
Frame = -3
Query: 343 KNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+N EEDEDE+ ++ DDDD D EDE +EDED D+
Sbjct: 460 ENGEEDEDEDGDDQDDDDEDDDDDDEEDEAGEEEDEDGAEDE 335
Score = 38.1 bits (87), Expect = 0.007
Identities = 20/63 (31%), Positives = 34/63 (53%), Gaps = 3/63 (4%)
Frame = -3
Query: 322 DKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEE---MDDDDNVDFGTYTEDEDAIDEDE 378
+K + GG G ++++ED D++D++ DDDD D EDED +++E
Sbjct: 511 NKSNSKKAPEGGAGGAEENGEEDEDEDGDDQDDDDEDDDDDDEEDEAGEEEDEDGAEDEE 332
Query: 379 DEE 381
+EE
Sbjct: 331 NEE 323
Score = 32.7 bits (73), Expect = 0.29
Identities = 17/45 (37%), Positives = 24/45 (52%)
Frame = -3
Query: 333 GETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDED 377
GE+ + S + E ED D+E DDDD+ D G +E+ ED
Sbjct: 673 GESKIQSEDKQRDAEGEDGNDDEGDDDDDGDGGFGEGEEELSSED 539
Score = 30.0 bits (66), Expect = 1.8
Identities = 11/39 (28%), Positives = 26/39 (66%)
Frame = -3
Query: 343 KNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
++ ++++D++D+E D+ + EDE+ +++EDEE
Sbjct: 421 QDDDDEDDDDDDEEDEAGEEEDEDGAEDEENEEDEEDEE 305
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 40.0 bits (92), Expect = 0.002
Identities = 22/80 (27%), Positives = 42/80 (52%)
Frame = +2
Query: 304 KRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDDNVD 363
K + K+L E+ E + + E+ V+ +++E+ED++D+E DD+D+ +
Sbjct: 218 KMKATKELSEQVEENEVKVLVEEDDEEYESVDEVVDDAEDDEDEEEDDDDDEGDDNDDEE 397
Query: 364 FGTYTEDEDAIDEDEDEEVD 383
+D DED++EE D
Sbjct: 398 DDAPDGGDDDDDEDDEEEGD 457
Score = 37.7 bits (86), Expect = 0.009
Identities = 30/102 (29%), Positives = 47/102 (45%), Gaps = 7/102 (6%)
Frame = +2
Query: 287 VEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKE 346
VE EEY E+ +D +++ E++ E D E + G + +
Sbjct: 278 VEEDDEEY-ESVDEVVDDAEDDEDEEEDDDDDEGDDNDDEEDDAPDGG---------DDD 427
Query: 347 EDEDEEDE-------EMDDDDNVDFGTYTEDEDAIDEDEDEE 381
+DED+E+E E DDDDN +DED +EDE+E+
Sbjct: 428 DDEDDEEEGDVQRGGEPDDDDNDSDDDSDDDEDEDEEDEEEQ 553
Score = 29.6 bits (65), Expect = 2.4
Identities = 15/36 (41%), Positives = 20/36 (54%)
Frame = +2
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
E D+D+ D + D DD+ EDED DE+E E
Sbjct: 473 EPDDDDNDSDDDSDDD-------EDEDEEDEEEQGE 559
Score = 29.3 bits (64), Expect = 3.2
Identities = 18/44 (40%), Positives = 24/44 (53%), Gaps = 8/44 (18%)
Frame = +2
Query: 346 EEDEDEEDEEMDDDDNVDFGT--------YTEDEDAIDEDEDEE 381
+EDEDEEDEE + + D GT E+E+A + E EE
Sbjct: 518 DEDEDEEDEE-EQGEEEDLGTEYLIRPLETAEEEEASSDFEPEE 646
>TC213554 weakly similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1,
partial (18%)
Length = 451
Score = 39.7 bits (91), Expect = 0.002
Identities = 24/73 (32%), Positives = 40/73 (53%)
Frame = +2
Query: 307 EEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGT 366
++ Q EN +E +++ E+E+ E V + +EE+EDEEDEE +++D D
Sbjct: 149 KQHQPKPENAAQEYEEEVEEEVEEEEVEEEVE----EEEEEEEDEEDEEEEEEDQKD--Q 310
Query: 367 YTEDEDAIDEDED 379
Y + DED+D
Sbjct: 311 YQHQQQQQDEDDD 349
Score = 29.6 bits (65), Expect = 2.4
Identities = 14/40 (35%), Positives = 26/40 (65%)
Frame = +2
Query: 345 KEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+E +E+E +EE+++++ E+ED DE+E+EE K
Sbjct: 206 EEVEEEEVEEEVEEEEE-------EEEDEEDEEEEEEDQK 304
>TC224567 similar to PIR|T51159|T51159 HMG protein [imported] - Arabidopsis
thaliana {Arabidopsis thaliana;} , partial (71%)
Length = 876
Score = 39.7 bits (91), Expect = 0.002
Identities = 35/127 (27%), Positives = 59/127 (45%), Gaps = 5/127 (3%)
Frame = +2
Query: 242 ASSDQSYTSNLTQSLSLALDEFYSNLRSAG--VSAVTGAGIEGFFKAVEASAEEYMETYK 299
A D + + + L+EF ++ V AV+ G G K S+ E Y+
Sbjct: 263 AKKDPNKPKRPPSAFFVFLEEFRKTFKAENPLVKAVSVVGKAGGEKWKSLSSAE-KAPYE 439
Query: 300 ADLDKRREEKQLL---EENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEM 356
A KR+ E + L E ++ + D + KS ++G ++ +EDED+E+EE
Sbjct: 440 AKAAKRKAEYEKLIKAYEKKQASSADDDESDKSKSEVNDEDDASGEEDHQEDEDDEEEED 619
Query: 357 DDDDNVD 363
D+DD+ D
Sbjct: 620 DEDDD*D 640
Score = 35.4 bits (80), Expect = 0.044
Identities = 21/68 (30%), Positives = 34/68 (49%)
Frame = +2
Query: 316 RRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAID 375
+RK +KL + EK + K+K E DE+D ++D + EDED +
Sbjct: 452 KRKAEYEKLIKAYEKKQASSADDDESDKSKSEVNDEDDASGEED-------HQEDEDDEE 610
Query: 376 EDEDEEVD 383
E++DE+ D
Sbjct: 611 EEDDEDDD 634
>TC215450 similar to UP|P93488 (P93488) NAP1Ps, partial (30%)
Length = 672
Score = 38.9 bits (89), Expect = 0.004
Identities = 19/39 (48%), Positives = 26/39 (65%)
Frame = +3
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
E+DED+ED E DDD+ + EDED D+++DEE K
Sbjct: 210 EDDEDDEDIEEDDDEEEE-----EDEDDDDDEDDEEESK 311
Score = 29.6 bits (65), Expect = 2.4
Identities = 14/37 (37%), Positives = 22/37 (58%)
Frame = +3
Query: 345 KEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
+E+D++EE+E+ DDDD DED++EE
Sbjct: 237 EEDDDEEEEEDEDDDD--------------DEDDEEE 305
>TC215449 similar to UP|P93488 (P93488) NAP1Ps, partial (97%)
Length = 1495
Score = 38.9 bits (89), Expect = 0.004
Identities = 19/39 (48%), Positives = 26/39 (65%)
Frame = +2
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
E+DED+ED E DDD+ + EDED D+++DEE K
Sbjct: 1043 EDDEDDEDIEEDDDEEEE-----EDEDDDDDEDDEEESK 1144
Score = 29.6 bits (65), Expect = 2.4
Identities = 14/37 (37%), Positives = 22/37 (58%)
Frame = +2
Query: 345 KEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
+E+D++EE+E+ DDDD DED++EE
Sbjct: 1070 EEDDDEEEEEDEDDDD--------------DEDDEEE 1138
>TC224760 similar to UP|O04692 (O04692) DNA-binding protein, partial (73%)
Length = 654
Score = 38.9 bits (89), Expect = 0.004
Identities = 35/132 (26%), Positives = 62/132 (46%), Gaps = 5/132 (3%)
Frame = +3
Query: 237 VFQAAASSDQSYTSNLTQSLSLALDEFYSNLRSAG--VSAVTGAGIEGFFKAVEASAEEY 294
+F A + + + +T + +EF ++ V AV+ G G K S+ E
Sbjct: 66 IFFALSFISLPWKNGMTYFVLCDSEEFRKTFKAENPLVKAVSVVGKAGGEKWKSLSSAE- 242
Query: 295 METYKADLDKRREEKQLL---EENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDE 351
Y+A KR+ E + L E ++ + D + KS ++G ++ +EDED+
Sbjct: 243 KAPYEAKAAKRKAEYEKLIKAYEKKQASSADDDESDKSKSEVNDEDDASGEEDHQEDEDD 422
Query: 352 EDEEMDDDDNVD 363
E+EE D+DD+ D
Sbjct: 423 EEEEDDEDDD*D 458
Score = 35.4 bits (80), Expect = 0.044
Identities = 21/68 (30%), Positives = 34/68 (49%)
Frame = +3
Query: 316 RRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAID 375
+RK +KL + EK + K+K E DE+D ++D + EDED +
Sbjct: 270 KRKAEYEKLIKAYEKKQASSADDDESDKSKSEVNDEDDASGEED-------HQEDEDDEE 428
Query: 376 EDEDEEVD 383
E++DE+ D
Sbjct: 429 EEDDEDDD 452
>TC215452 similar to UP|P93488 (P93488) NAP1Ps, partial (29%)
Length = 609
Score = 38.5 bits (88), Expect = 0.005
Identities = 18/39 (46%), Positives = 27/39 (69%)
Frame = +3
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
E+DED+ED E DDD++ + +D+D D+D+DEE K
Sbjct: 138 EDDEDDEDIEEDDDEDEE-----DDDDDDDDDDDEEESK 239
Score = 30.4 bits (67), Expect = 1.4
Identities = 11/19 (57%), Positives = 17/19 (88%)
Frame = +3
Query: 345 KEEDEDEEDEEMDDDDNVD 363
+++DEDEED++ DDDD+ D
Sbjct: 168 EDDDEDEEDDDDDDDDDDD 224
>TC215451 weakly similar to UP|Q94K07 (Q94K07) Nucleosome assembly protein,
partial (20%)
Length = 777
Score = 38.5 bits (88), Expect = 0.005
Identities = 18/39 (46%), Positives = 27/39 (69%)
Frame = +2
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
E+DED+ED E DDD++ + +D+D D+D+DEE K
Sbjct: 89 EDDEDDEDIEEDDDEDEE-----DDDDDDDDDDDEEESK 190
Score = 30.4 bits (67), Expect = 1.4
Identities = 11/19 (57%), Positives = 17/19 (88%)
Frame = +2
Query: 345 KEEDEDEEDEEMDDDDNVD 363
+++DEDEED++ DDDD+ D
Sbjct: 119 EDDDEDEEDDDDDDDDDDD 175
>TC224564 similar to PIR|T51159|T51159 HMG protein [imported] - Arabidopsis
thaliana {Arabidopsis thaliana;} , partial (82%)
Length = 833
Score = 38.5 bits (88), Expect = 0.005
Identities = 36/125 (28%), Positives = 59/125 (46%), Gaps = 3/125 (2%)
Frame = +1
Query: 242 ASSDQSYTSNLTQSLSLALDEFYSNLRSAG--VSAVTGAGIEGFFKAVEASAEEYMETYK 299
A D + + + L+EF ++ V AV+ G G K S+ E Y+
Sbjct: 220 AKKDPNKPKRPPSAFFVFLEEFRKTFKAENPNVKAVSVVGKAGGEKWKSLSSAE-KAPYE 396
Query: 300 ADLDKRREE-KQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDD 358
A KR+ E ++L++ +K+ E +KS E V +EE+ED+E+EE D+
Sbjct: 397 AKAAKRKAEYEKLIKAYDKKQASSADDEESDKSKSE-VNDEDDASGEEEEEDDEEEEDDE 573
Query: 359 DDNVD 363
DD+ D
Sbjct: 574 DDD*D 588
>TC215879 similar to UP|IF5_PHAVU (P48724) Eukaryotic translation initiation
factor 5 (eIF-5), partial (64%)
Length = 1110
Score = 37.7 bits (86), Expect = 0.009
Identities = 27/82 (32%), Positives = 40/82 (47%), Gaps = 4/82 (4%)
Frame = +1
Query: 305 RREEKQLLEENR-RKENMDKLRREMEKSGGETV---VLSTGLKNKEEDEDEEDEEMDDDD 360
RR EK+ L+E E K+ +E++K G + ST K K D ED
Sbjct: 10 RRAEKERLKEGELADEEQKKVNKEVKKKGSSSSKDGTKSTSSKKKASGSD-EDRTSPTHS 186
Query: 361 NVDFGTYTEDEDAIDEDEDEEV 382
+D E E+A+DED+D++V
Sbjct: 187 QID-----EKEEALDEDDDDDV 237
>TC218918 UP|Q39892 (Q39892) Nucleosome assembly protein 1, complete
Length = 1507
Score = 37.4 bits (85), Expect = 0.012
Identities = 32/109 (29%), Positives = 52/109 (47%), Gaps = 7/109 (6%)
Frame = +1
Query: 283 FFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTG- 341
FFK E ++ AD+D+ E+ +N+ +++ D +K V TG
Sbjct: 820 FFKPPEVPEDD------ADIDEDLAEEL---QNQMEQDYDIGSTLRDKIIPHAVSWFTGE 972
Query: 342 ------LKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
++ E+DEDEE++E +D+D D DED DE+ED+ K
Sbjct: 973 AAQGDEFEDLEDDEDEEEDEDEDEDEED------DEDEDDEEEDDTKTK 1101
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.133 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,020,355
Number of Sequences: 63676
Number of extensions: 171747
Number of successful extensions: 13369
Number of sequences better than 10.0: 290
Number of HSP's better than 10.0 without gapping: 4119
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10038
length of query: 387
length of database: 12,639,632
effective HSP length: 99
effective length of query: 288
effective length of database: 6,335,708
effective search space: 1824683904
effective search space used: 1824683904
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC121237.18


