

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121234.11 - phase: 0 /pseudo
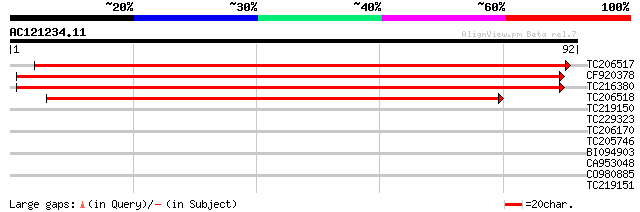
(92 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC206517 similar to UP|Q9XFI5 (Q9XFI5) NADPH:quinone oxidoreduct... 148 5e-37
CF920378 133 1e-32
TC216380 similar to UP|Q9LK88 (Q9LK88) NADPH:quinone oxidoreduct... 130 1e-31
TC206518 similar to UP|Q9LK88 (Q9LK88) NADPH:quinone oxidoreduct... 114 8e-27
TC219150 26 2.9
TC229323 similar to UP|Q6NXD3 (Q6NXD3) Zgc:77493 protein, partia... 25 5.0
TC206170 UP|CAP1_SOYBN (Q02909) Phosphoenolpyruvate carboxylase,... 25 5.0
TC205746 similar to UP|Q9FIJ4 (Q9FIJ4) Gb|AAF26028.1, partial (34%) 25 5.0
BI094903 25 6.5
CA953048 25 6.5
CO980885 25 8.5
TC219151 25 8.5
>TC206517 similar to UP|Q9XFI5 (Q9XFI5) NADPH:quinone oxidoreductase, partial
(95%)
Length = 771
Score = 148 bits (373), Expect = 5e-37
Identities = 75/87 (86%), Positives = 79/87 (90%)
Frame = +1
Query: 5 VIKVAALSGSIRKVSYHSGLIRAAIELSKGATEGIEIEFIDISTLPMYNTDLENEGTYPP 64
VIKVAALSGS+RK SYHSGLIRAAIELSKG EGIEIEFI+IS LPM NTDLE GTYPP
Sbjct: 58 VIKVAALSGSLRKASYHSGLIRAAIELSKGEIEGIEIEFIEISELPMLNTDLEINGTYPP 237
Query: 65 LVEAFRHKILQADSVLFASPEYNYSVT 91
+VEAFR KILQADS+LFASPEYNYSVT
Sbjct: 238 IVEAFRQKILQADSILFASPEYNYSVT 318
>CF920378
Length = 735
Score = 133 bits (335), Expect = 1e-32
Identities = 66/89 (74%), Positives = 78/89 (87%)
Frame = +3
Query: 2 ATAVIKVAALSGSIRKVSYHSGLIRAAIELSKGATEGIEIEFIDISTLPMYNTDLENEGT 61
+++VIKVAALSGS+RK SY+ GLIR+AIELSKG EG++IE++DIS LP+ NTDLE GT
Sbjct: 72 SSSVIKVAALSGSLRKGSYNRGLIRSAIELSKGRVEGLQIEYVDISPLPLLNTDLEVNGT 251
Query: 62 YPPLVEAFRHKILQADSVLFASPEYNYSV 90
YPP VEAFR KIL ADS+LFASPEYNYSV
Sbjct: 252 YPPEVEAFRQKILAADSILFASPEYNYSV 338
>TC216380 similar to UP|Q9LK88 (Q9LK88) NADPH:quinone oxidoreductase, partial
(95%)
Length = 998
Score = 130 bits (326), Expect = 1e-31
Identities = 64/89 (71%), Positives = 77/89 (85%)
Frame = +3
Query: 2 ATAVIKVAALSGSIRKVSYHSGLIRAAIELSKGATEGIEIEFIDISTLPMYNTDLENEGT 61
+++VIKVAALSGS+RK SY+ LIR+AIELS+G EG++IE++DIS LP+ NTDLE GT
Sbjct: 186 SSSVIKVAALSGSLRKGSYNRALIRSAIELSQGRVEGLQIEYVDISPLPLLNTDLEVNGT 365
Query: 62 YPPLVEAFRHKILQADSVLFASPEYNYSV 90
YPP VEAFR KIL ADS+LFASPEYNYSV
Sbjct: 366 YPPQVEAFRQKILAADSILFASPEYNYSV 452
>TC206518 similar to UP|Q9LK88 (Q9LK88) NADPH:quinone oxidoreductase,
partial (63%)
Length = 405
Score = 114 bits (285), Expect = 8e-27
Identities = 59/74 (79%), Positives = 64/74 (85%)
Frame = +1
Query: 7 KVAALSGSIRKVSYHSGLIRAAIELSKGATEGIEIEFIDISTLPMYNTDLENEGTYPPLV 66
KVAALSGS++K S H+GLIRAAIELSKGA EGIEIEFI+IS LPM NTDLEN GTYPP+V
Sbjct: 4 KVAALSGSLKKTSSHTGLIRAAIELSKGAIEGIEIEFIEISELPMLNTDLENNGTYPPIV 183
Query: 67 EAFRHKILQADSVL 80
EAFR KILQ S L
Sbjct: 184EAFRCKILQTHSGL 225
Score = 27.3 bits (59), Expect = 1.3
Identities = 12/26 (46%), Positives = 17/26 (65%)
Frame = +2
Query: 66 VEAFRHKILQADSVLFASPEYNYSVT 91
++ F + + V FASPEYNYS+T
Sbjct: 182 LKLFAARFFKLIVVSFASPEYNYSLT 259
>TC219150
Length = 655
Score = 26.2 bits (56), Expect = 2.9
Identities = 15/39 (38%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Frame = -1
Query: 52 YNTDLENEGTYPPLVEAFRHKILQADSVLFAS--PEYNY 88
+++D+E E TYP +V R K L A + AS P+Y +
Sbjct: 259 FSSDIEVEHTYPTIVNINRTKPLWAQA*SSAS*CPQYGF 143
>TC229323 similar to UP|Q6NXD3 (Q6NXD3) Zgc:77493 protein, partial (42%)
Length = 1362
Score = 25.4 bits (54), Expect = 5.0
Identities = 19/64 (29%), Positives = 28/64 (43%)
Frame = +3
Query: 15 IRKVSYHSGLIRAAIELSKGATEGIEIEFIDISTLPMYNTDLENEGTYPPLVEAFRHKIL 74
+R +S + A+EL E + E ID L++ GTY L +HK+L
Sbjct: 114 LRYLSKDDFRVLTAVELGMRNHEIVPTELIDRIAR------LKHGGTYKVLKNLLKHKLL 275
Query: 75 QADS 78
DS
Sbjct: 276 HHDS 287
>TC206170 UP|CAP1_SOYBN (Q02909) Phosphoenolpyruvate carboxylase, housekeeping
isozyme (PEPCase) , complete
Length = 3286
Score = 25.4 bits (54), Expect = 5.0
Identities = 10/17 (58%), Positives = 12/17 (69%)
Frame = +1
Query: 62 YPPLVEAFRHKILQADS 78
+PPL F +KILQ DS
Sbjct: 3001 FPPLYPCFHYKILQRDS 3051
>TC205746 similar to UP|Q9FIJ4 (Q9FIJ4) Gb|AAF26028.1, partial (34%)
Length = 521
Score = 25.4 bits (54), Expect = 5.0
Identities = 14/44 (31%), Positives = 23/44 (51%)
Frame = -2
Query: 47 STLPMYNTDLENEGTYPPLVEAFRHKILQADSVLFASPEYNYSV 90
S+ P Y T L PPL + ++K L+A ++ +S Y Y +
Sbjct: 388 SSFPQYLTVLYRST*QPPLFTSNQNKNLEA*KLVISSS*YKYKL 257
>BI094903
Length = 419
Score = 25.0 bits (53), Expect = 6.5
Identities = 13/33 (39%), Positives = 19/33 (57%)
Frame = +1
Query: 6 IKVAALSGSIRKVSYHSGLIRAAIELSKGATEG 38
IK +LSGS++K S+ +ELS+ A G
Sbjct: 97 IKANSLSGSLQKQSFEGMKQLEVVELSENALTG 195
>CA953048
Length = 427
Score = 25.0 bits (53), Expect = 6.5
Identities = 11/33 (33%), Positives = 17/33 (51%)
Frame = -1
Query: 48 TLPMYNTDLENEGTYPPLVEAFRHKILQADSVL 80
TLP ++EN+ + P ++EA I Q L
Sbjct: 232 TLPQQGMEIENQVSLPHMLEALGEAIYQGKKAL 134
>CO980885
Length = 759
Score = 24.6 bits (52), Expect = 8.5
Identities = 10/20 (50%), Positives = 16/20 (80%)
Frame = -1
Query: 66 VEAFRHKILQADSVLFASPE 85
V AF HK+L++ ++L +SPE
Sbjct: 534 VSAFFHKLLESSTLL*SSPE 475
>TC219151
Length = 759
Score = 24.6 bits (52), Expect = 8.5
Identities = 15/39 (38%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Frame = -3
Query: 52 YNTDLENEGTYPPLVEAFRHKILQADSVLFAS--PEYNY 88
+++D+E E TYP +V R K L A + AS P+Y +
Sbjct: 349 FSSDIEVEYTYPTIVNINRSKPLWA*A*SSAS*CPQYRF 233
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.133 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,846,133
Number of Sequences: 63676
Number of extensions: 25379
Number of successful extensions: 103
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 103
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 103
length of query: 92
length of database: 12,639,632
effective HSP length: 68
effective length of query: 24
effective length of database: 8,309,664
effective search space: 199431936
effective search space used: 199431936
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 51 (24.3 bits)
Medicago: description of AC121234.11


