

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC119408.7 + phase: 0 /pseudo
(218 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
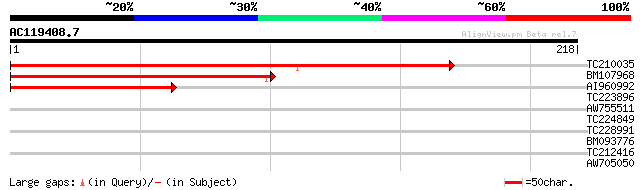
Score E
Sequences producing significant alignments: (bits) Value
TC210035 296 5e-81
BM107968 172 1e-43
AI960992 125 2e-29
TC223896 UP|Q7FPX7 (Q7FPX7) Polygalacturonase inhibitor protein,... 31 0.50
AW755511 similar to GP|11120514|gb| cytokinin oxidase {Arabidops... 28 2.5
TC224849 similar to GB|AAN46773.1|24111299|BT001019 At3g52990/F8... 28 3.2
TC228991 similar to GB|AAQ22666.1|33589800|BT010197 At3g63530 {A... 27 5.5
BM093776 27 7.2
TC212416 homologue to UP|Q89QR3 (Q89QR3) Bll3061 protein, partia... 27 7.2
AW705050 27 9.4
>TC210035
Length = 927
Score = 296 bits (758), Expect = 5e-81
Identities = 157/221 (71%), Positives = 162/221 (73%), Gaps = 50/221 (22%)
Frame = +2
Query: 1 MAAPFFSTPFQPYVYQSQQDAIIPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVE 60
MAAPFFSTPFQPYVYQS QDAIIPFQILGGE+QVVQIMLKP EKIIAKPGSMCFMSGS+E
Sbjct: 221 MAAPFFSTPFQPYVYQSPQDAIIPFQILGGEAQVVQIMLKPQEKIIAKPGSMCFMSGSIE 400
Query: 61 MENAYLPENEVGIWQWLFGKTVTNIVVRNSGPSDGFVGIAAPYFARILP----------- 109
MENAYLPENEVGIWQWLFGKT+T+IV+ NSGPSDGFVGIAAPYFARILP
Sbjct: 401 MENAYLPENEVGIWQWLFGKTITSIVLCNSGPSDGFVGIAAPYFARILPVCTSMPLQLS* 580
Query: 110 ---------------------------------------IDLATFNGEILCQPDAFLCSV 130
IDLA FNGEILCQPDAFLCSV
Sbjct: 581 IIHCYSCFKSRY**SETHICIHLTEFLFIFGYVSVTQIQIDLAMFNGEILCQPDAFLCSV 760
Query: 131 NDVKVSNTVDQRGRNVVAGAEVFLRQKLSGQGLAFILGGGS 171
NDVKVSN VDQRGRN VA AE FLRQKLSGQGLAFIL GGS
Sbjct: 761 NDVKVSNIVDQRGRNAVASAEGFLRQKLSGQGLAFILAGGS 883
>BM107968
Length = 371
Score = 172 bits (435), Expect = 1e-43
Identities = 86/103 (83%), Positives = 90/103 (86%), Gaps = 1/103 (0%)
Frame = +3
Query: 1 MAAPFFSTPFQPYVYQSQQDAIIPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVE 60
MAAPFFSTPFQPYVY S QDAIIPFQILGGE+QVVQIMLKP EKIIAKPGSMCFMSGSVE
Sbjct: 63 MAAPFFSTPFQPYVYXSPQDAIIPFQILGGEAQVVQIMLKPQEKIIAKPGSMCFMSGSVE 242
Query: 61 MENAYLPENEVGIWQWLFGKTVTNIVVRNSGPSDGFV-GIAAP 102
MENAYLPENE W WL GKT+T IV+RNSGPS + GIAAP
Sbjct: 243 MENAYLPENEXXXWXWLXGKTITXIVLRNSGPS*WILFGIAAP 371
>AI960992
Length = 434
Score = 125 bits (313), Expect = 2e-29
Identities = 60/64 (93%), Positives = 62/64 (96%)
Frame = +2
Query: 1 MAAPFFSTPFQPYVYQSQQDAIIPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVE 60
MAAPFFSTPFQPYVYQS QDAIIPFQILGGE+QVVQIMLKP EKIIAKPGSMCFMSGS+E
Sbjct: 242 MAAPFFSTPFQPYVYQSPQDAIIPFQILGGEAQVVQIMLKPQEKIIAKPGSMCFMSGSIE 421
Query: 61 MENA 64
MENA
Sbjct: 422 MENA 433
>TC223896 UP|Q7FPX7 (Q7FPX7) Polygalacturonase inhibitor protein, complete
Length = 1094
Score = 30.8 bits (68), Expect = 0.50
Identities = 27/80 (33%), Positives = 37/80 (45%), Gaps = 2/80 (2%)
Frame = -3
Query: 121 CQPDAFLCSVNDVKVSNTVDQRGRNVVAGAEVFLRQ-KLSGQGLAFILGGGSVVQKILEV 179
C D +C V ++V D RG EV + + K+ G GG V ++V
Sbjct: 399 CARDIGVCDVEIIEVGEISDGRGD---CAEEVCVNEVKIIQVGKVAYGGGNRVGFGEVKV 229
Query: 180 GEVLAVDVSC-IVAVTSTVD 198
GEV VD +C +VAV T D
Sbjct: 228 GEVEDVDAACLVVAVGDTSD 169
>AW755511 similar to GP|11120514|gb| cytokinin oxidase {Arabidopsis
thaliana}, partial (12%)
Length = 433
Score = 28.5 bits (62), Expect = 2.5
Identities = 20/65 (30%), Positives = 33/65 (50%), Gaps = 3/65 (4%)
Frame = -3
Query: 132 DVKVSNTVDQRGRNVVAGAEVFLRQKLSGQGLAFIL---GGGSVVQKILEVGEVLAVDVS 188
+V+++ R R+V RQ+LSG A +L GGGS V+ +E V+
Sbjct: 311 EVRLARRRLHRSRHVAGDGGANNRQRLSGFHPAEVLAGSGGGSEVESSVESRNVVVWHRF 132
Query: 189 CIVAV 193
C+++V
Sbjct: 131 CVISV 117
>TC224849 similar to GB|AAN46773.1|24111299|BT001019 At3g52990/F8J2_160
{Arabidopsis thaliana;} , complete
Length = 1966
Score = 28.1 bits (61), Expect = 3.2
Identities = 22/82 (26%), Positives = 36/82 (43%)
Frame = +1
Query: 116 NGEILCQPDAFLCSVNDVKVSNTVDQRGRNVVAGAEVFLRQKLSGQGLAFILGGGSVVQK 175
NG ++ PD + +++ N D + V G +F+ Q ++ G
Sbjct: 454 NGLVVLTPDQGQEASSEILPIN-FDGLAKAVKKGDTIFIGQ--------YLFTGSETTSV 606
Query: 176 ILEVGEVLAVDVSCIVAVTSTV 197
LEV EV DV CI+ T+T+
Sbjct: 607 WLEVSEVKGQDVVCIIKNTATL 672
>TC228991 similar to GB|AAQ22666.1|33589800|BT010197 At3g63530 {Arabidopsis
thaliana;} , partial (46%)
Length = 1167
Score = 27.3 bits (59), Expect = 5.5
Identities = 16/55 (29%), Positives = 30/55 (54%)
Frame = +3
Query: 31 ESQVVQIMLKPHEKIIAKPGSMCFMSGSVEMENAYLPENEVGIWQWLFGKTVTNI 85
ES + + P++ ++ PGS + S S E+ N +LP E+ +W + T+T +
Sbjct: 288 ESVYWSMNMNPYKFGLSGPGSTSYYS-SYEV-NGHLPRMEIDRAEWEYPSTITTV 446
>BM093776
Length = 381
Score = 26.9 bits (58), Expect = 7.2
Identities = 14/42 (33%), Positives = 19/42 (44%)
Frame = +2
Query: 49 PGSMCFMSGSVEMENAYLPENEVGIWQWLFGKTVTNIVVRNS 90
P ++CF P V W WL GK VTN+ + +S
Sbjct: 218 PSTLCFF--------LLFPSLLVN*WSWLIGK*VTNVCLFSS 319
>TC212416 homologue to UP|Q89QR3 (Q89QR3) Bll3061 protein, partial (60%)
Length = 276
Score = 26.9 bits (58), Expect = 7.2
Identities = 12/38 (31%), Positives = 22/38 (57%)
Frame = +2
Query: 145 NVVAGAEVFLRQKLSGQGLAFILGGGSVVQKILEVGEV 182
N + GAE + SG GL+ ++GGG + +I + ++
Sbjct: 161 NHIPGAEAAIEASKSGGGLSRLMGGGRMA*RITKEEQI 274
>AW705050
Length = 365
Score = 26.6 bits (57), Expect = 9.4
Identities = 11/28 (39%), Positives = 16/28 (56%)
Frame = +1
Query: 109 PIDLATFNGEILCQPDAFLCSVNDVKVS 136
P+ FNG+++C LC+ N V VS
Sbjct: 109 PLQFPGFNGKLICPAYHELCNTNPVVVS 192
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.139 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,864,640
Number of Sequences: 63676
Number of extensions: 106253
Number of successful extensions: 451
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 450
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 451
length of query: 218
length of database: 12,639,632
effective HSP length: 93
effective length of query: 125
effective length of database: 6,717,764
effective search space: 839720500
effective search space used: 839720500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC119408.7


