
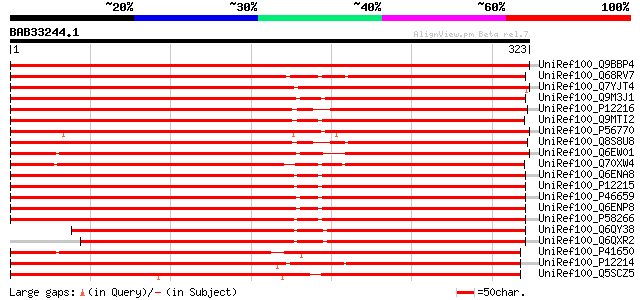
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BAB33244.1 323 aa
(323 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9BBP4 Cytochrome c biogenesis protein ccsA [Lotus jap... 654 0.0
UniRef100_Q68RV7 Hypothetical protein ccsA [Panax ginseng] 491 e-138
UniRef100_Q7YJT4 Ycf5 protein [Calycanthus fertilis var. ferax] 484 e-135
UniRef100_Q9M3J1 Cytochrome c biogenesis protein ccsA [Spinacia ... 482 e-135
UniRef100_P12216 Cytochrome c biogenesis protein ccsA [Nicotiana... 474 e-132
UniRef100_Q9MTI2 Cytochrome c biogenesis protein ccsA [Oenothera... 471 e-132
UniRef100_P56770 Cytochrome c biogenesis protein ccsA [Arabidops... 467 e-130
UniRef100_Q8S8U8 CcsA protein [Atropa belladonna] 463 e-129
UniRef100_Q6EW01 CcsA protein [Nymphaea alba] 461 e-128
UniRef100_Q70XW4 CcsA protein [Amborella trichopoda] 457 e-127
UniRef100_Q6ENA8 Heme attachment protein [Oryza nivara] 451 e-125
UniRef100_P12215 Cytochrome c biogenesis protein ccsA [Oryza sat... 451 e-125
UniRef100_P46659 Cytochrome c biogenesis protein ccsA [Zea mays] 447 e-124
UniRef100_Q6ENP8 C-type cytochrome synthesis [Saccharum officina... 446 e-124
UniRef100_P58266 Cytochrome c biogenesis protein ccsA [Triticum ... 432 e-120
UniRef100_Q6QY38 Cytochrome c biogenesis protein [Oryza sativa] 400 e-110
UniRef100_Q6QXR2 Cytochrome c biogenesis protein [Oryza sativa] 389 e-107
UniRef100_P41650 Cytochrome c biogenesis protein ccsA [Pinus thu... 344 2e-93
UniRef100_P12214 Cytochrome c biogenesis protein ccsA [Marchanti... 317 3e-85
UniRef100_Q5SCZ5 Cytochrome c heme attachment protein [Huperzia ... 310 3e-83
>UniRef100_Q9BBP4 Cytochrome c biogenesis protein ccsA [Lotus japonicus]
Length = 323
Score = 654 bits (1688), Expect = 0.0
Identities = 323/323 (100%), Positives = 323/323 (100%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW
Sbjct: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAPSVIFTQGFATSGLLTE 120
FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAPSVIFTQGFATSGLLTE
Sbjct: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAPSVIFTQGFATSGLLTE 120
Query: 121 MHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKSKRLSF 180
MHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKSKRLSF
Sbjct: 121 MHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKSKRLSF 180
Query: 181 LYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLTIG 240
LYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLTIG
Sbjct: 181 LYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLTIG 240
Query: 241 ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFLII 300
ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFLII
Sbjct: 241 ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFLII 300
Query: 301 WICYFGVNLLGIGLHNYGSFTSN 323
WICYFGVNLLGIGLHNYGSFTSN
Sbjct: 301 WICYFGVNLLGIGLHNYGSFTSN 323
>UniRef100_Q68RV7 Hypothetical protein ccsA [Panax ginseng]
Length = 320
Score = 491 bits (1265), Expect = e-138
Identities = 247/321 (76%), Positives = 274/321 (84%), Gaps = 5/321 (1%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
MIFSTLEHILTHISFS+VSIVI+IHLITL V++I+ YDSS+KGMI FLCITGLL+TRW
Sbjct: 1 MIFSTLEHILTHISFSIVSIVITIHLITLLVDEIIKLYDSSEKGMIAIFLCITGLLVTRW 60
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAPSVIFTQGFATSGLLTE 120
+S H P SDLYESLIFLSW S+ ++VP FKK+K +LSTI A SVIFTQGFATSGLLTE
Sbjct: 61 IYSRHFPLSDLYESLIFLSWSLSVIHIVPYFKKKKKNLSTITASSVIFTQGFATSGLLTE 120
Query: 121 MHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKSKRLSF 180
+HQS ILVPALQS WL+MHVSMMIL YAALLCGSLLSVA+LVITF++ I KR
Sbjct: 121 IHQSAILVPALQSEWLIMHVSMMILSYAALLCGSLLSVALLVITFRKNRNIF--CKRNPL 178
Query: 181 LYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLTIG 240
L E F + EI+Y MNERNNVLR T FS+ +NYYR Q IQQLD W YR+ISLGFIFLTIG
Sbjct: 179 LNELFSFGEIQY--MNERNNVLRTTFFSA-KNYYRSQLIQQLDYWSYRVISLGFIFLTIG 235
Query: 241 ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFLII 300
ILSGAVWANEAWGSYWNWDPKETWAFITW +FAIYLH+R N KL G NS+IVASIGFLII
Sbjct: 236 ILSGAVWANEAWGSYWNWDPKETWAFITWIVFAIYLHTRTNIKLRGANSAIVASIGFLII 295
Query: 301 WICYFGVNLLGIGLHNYGSFT 321
WICYFGVNLLGIGLH+YGSFT
Sbjct: 296 WICYFGVNLLGIGLHSYGSFT 316
>UniRef100_Q7YJT4 Ycf5 protein [Calycanthus fertilis var. ferax]
Length = 323
Score = 484 bits (1247), Expect = e-135
Identities = 236/325 (72%), Positives = 271/325 (82%), Gaps = 4/325 (1%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
MIFSTLEHILTHISFS++SIVI+IHL+ L V+ VG DSS+KGM+ TF CITGLL+TRW
Sbjct: 1 MIFSTLEHILTHISFSIISIVITIHLLNLLVHDTVGLCDSSEKGMMATFFCITGLLVTRW 60
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAPSVIFTQGFATSGLLTE 120
+S H P SDLYESL+FLSW FSI +MVP F+ KN LS I APS IFTQGFATSGLLTE
Sbjct: 61 IYSRHFPLSDLYESLMFLSWSFSIIHMVPYFRNHKNSLSAITAPSAIFTQGFATSGLLTE 120
Query: 121 MHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKSKRLSF 180
MHQS ILVPALQS WLMMHVSMM+LGYA+LLCGSLLSVA+LVITF++ + I GK+ F
Sbjct: 121 MHQSAILVPALQSQWLMMHVSMMLLGYASLLCGSLLSVALLVITFRKTLGIPGKNNH--F 178
Query: 181 LYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLTIG 240
L SF + EI+Y N + +VL+KTSF S+RNY+RYQ Q+LD YRIISLGF F TIG
Sbjct: 179 LIGSFPFGEIRYFNEKRKRSVLKKTSFFSFRNYHRYQLTQRLDHCSYRIISLGFTFSTIG 238
Query: 241 ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFLII 300
ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLH+R N++ + + +IVAS+GFLII
Sbjct: 239 ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHTRTNQRFQDVGPAIVASMGFLII 298
Query: 301 WICYFGVNLLGIGLHNYGSF--TSN 323
WICYFGVNLLGIGLH+YGSF TSN
Sbjct: 299 WICYFGVNLLGIGLHSYGSFALTSN 323
>UniRef100_Q9M3J1 Cytochrome c biogenesis protein ccsA [Spinacia oleracea]
Length = 323
Score = 482 bits (1241), Expect = e-135
Identities = 240/323 (74%), Positives = 271/323 (83%), Gaps = 6/323 (1%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
MIFSTLEHILTHISFS VS+VI++HLITL VN+IVG Y+S +KGM++TF CITGLL+TRW
Sbjct: 1 MIFSTLEHILTHISFSTVSVVITLHLITLLVNEIVGLYNSLEKGMLVTFFCITGLLVTRW 60
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRF-KKQKNDLSTIIAPSVIFTQGFATSGLLT 119
+ H P SDLYESLIFLSW F + +M+P F KK+KN L+ I APS IFTQGFATSGLLT
Sbjct: 61 VYWKHFPLSDLYESLIFLSWSFYLIHMIPSFLKKEKNSLNVITAPSAIFTQGFATSGLLT 120
Query: 120 EMHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQ-ELIPILGKSKRL 178
EMHQS ILVPALQS WLMMHVSMM+LGYAALL GSLLSV +L+I FQ +LI + K K L
Sbjct: 121 EMHQSGILVPALQSQWLMMHVSMMVLGYAALLGGSLLSVTLLIIIFQKDLIQVFDKRKHL 180
Query: 179 SFLYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLT 238
L ESF + EI+YIN E++N+++ S S RNYYR Q I+QLD W YR+ISLGFIFLT
Sbjct: 181 --LNESFFFGEIQYIN--EKSNIVQNASPSYVRNYYRSQLIEQLDHWSYRVISLGFIFLT 236
Query: 239 IGILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFL 298
IGILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLH R NK L G NS+IVA IGFL
Sbjct: 237 IGILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHIRTNKNLRGANSAIVAFIGFL 296
Query: 299 IIWICYFGVNLLGIGLHNYGSFT 321
IIWICYFGVNLLGIGLH+YGSFT
Sbjct: 297 IIWICYFGVNLLGIGLHSYGSFT 319
>UniRef100_P12216 Cytochrome c biogenesis protein ccsA [Nicotiana tabacum]
Length = 313
Score = 474 bits (1219), Expect = e-132
Identities = 237/322 (73%), Positives = 264/322 (81%), Gaps = 12/322 (3%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
MIFSTLEHILTHISFS+VSIVI+IHLIT V++IV YDSS+KG+I+TF CITGLL+TRW
Sbjct: 1 MIFSTLEHILTHISFSIVSIVITIHLITFLVDEIVKLYDSSEKGIIVTFFCITGLLVTRW 60
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAPSVIFTQGFATSGLLTE 120
SGH P SDLYESLIFLSW FS+ +++P FKK LS I PS IFTQGFATSG+LTE
Sbjct: 61 ISSGHFPLSDLYESLIFLSWSFSLIHIIPYFKKNVLILSKITGPSAIFTQGFATSGILTE 120
Query: 121 MHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKSKRLSF 180
+HQSVILVPALQS WL+MHVSMMILGYAALLCGSLLSVA+LVITF++ + KS F
Sbjct: 121 IHQSVILVPALQSEWLIMHVSMMILGYAALLCGSLLSVALLVITFRKNRQLFYKSN--GF 178
Query: 181 LYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLTIG 240
L ESF E NVL+ TSF S +NYYR Q IQQLD W YR+ISLGF FLTIG
Sbjct: 179 LNESFFLGE----------NVLQNTSFFSAKNYYRSQLIQQLDYWSYRVISLGFTFLTIG 228
Query: 241 ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFLII 300
ILSGAVWANEAWGSYWNWDPKETWAFITW +FAIYLH+R N+ L G NS+IVASIGFLII
Sbjct: 229 ILSGAVWANEAWGSYWNWDPKETWAFITWIVFAIYLHTRTNRNLRGANSAIVASIGFLII 288
Query: 301 WICYFGVNLLGIGLHNYGSFTS 322
WICYFGVNLLGIGLH+YGSF S
Sbjct: 289 WICYFGVNLLGIGLHSYGSFPS 310
>UniRef100_Q9MTI2 Cytochrome c biogenesis protein ccsA [Oenothera hookeri]
Length = 319
Score = 471 bits (1213), Expect = e-132
Identities = 233/320 (72%), Positives = 269/320 (83%), Gaps = 4/320 (1%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
MIF TLEHILTHISFS+VSI I+I LITL V++I+G YDSS+KG+I TFLCITGLL+TRW
Sbjct: 1 MIFYTLEHILTHISFSLVSIGITIFLITLSVDEIIGLYDSSEKGVIGTFLCITGLLVTRW 60
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAPSVIFTQGFATSGLLTE 120
+SGH P S+LYESL+FLSW F+I +M P FKKQK+ + TI + S IFTQG TSGLL+E
Sbjct: 61 AYSGHFPLSNLYESLLFLSWSFAIIHMFPYFKKQKSYVRTITSSSTIFTQGLVTSGLLSE 120
Query: 121 MHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKSKRLSF 180
M QS ILVPALQS WLMMHVSMM+LGYAALLCGSLLSVA+LVITF++ + I K K +F
Sbjct: 121 MQQSEILVPALQSQWLMMHVSMMVLGYAALLCGSLLSVALLVITFRKALRIFSKKK--AF 178
Query: 181 LYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLTIG 240
L +SF + EI+Y NE +NVL TSF S +NYYR Q IQQLDRW RIISLGFIFLTIG
Sbjct: 179 LKDSFSFVEIQY--RNEPSNVLLSTSFISSKNYYRAQLIQQLDRWSSRIISLGFIFLTIG 236
Query: 241 ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFLII 300
ILSGAVWANEAWGSYWNWDPKETWAFITWT+FAIYLH+R N + +NS+IVA +GF+II
Sbjct: 237 ILSGAVWANEAWGSYWNWDPKETWAFITWTMFAIYLHTRTNPNFQSVNSAIVAFLGFIII 296
Query: 301 WICYFGVNLLGIGLHNYGSF 320
WICYFGVNLLGIGLH+YGSF
Sbjct: 297 WICYFGVNLLGIGLHSYGSF 316
>UniRef100_P56770 Cytochrome c biogenesis protein ccsA [Arabidopsis thaliana]
Length = 328
Score = 467 bits (1201), Expect = e-130
Identities = 236/330 (71%), Positives = 273/330 (82%), Gaps = 9/330 (2%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVN--QIVGFYDSSKKGMIITFLCITGLLIT 58
MIFS LEHILTHISFSVVSIV++I+ +TL VN +I+GF+DSS KG+IITF ITGLL+T
Sbjct: 1 MIFSILEHILTHISFSVVSIVLTIYFLTLLVNLDEIIGFFDSSDKGIIITFFGITGLLLT 60
Query: 59 RWFFSGHLPFSDLYESLIFLSWGFSIFYMVPRF-KKQKNDLSTIIAPSVIFTQGFATSGL 117
RW +SGH P S+LYESLIFLSW FSI +MV F KKQ+N L+TI APSVIF QGFATSGL
Sbjct: 61 RWIYSGHFPLSNLYESLIFLSWAFSIIHMVSYFNKKQQNKLNTITAPSVIFIQGFATSGL 120
Query: 118 LTEMHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKS-- 175
L +M QS ILVPALQS WLMMHVSMMILGY ALLCGSLLS+A+LVITF+++ P K
Sbjct: 121 LNKMPQSAILVPALQSQWLMMHVSMMILGYGALLCGSLLSIALLVITFRKVGPTFWKKNI 180
Query: 176 KRLSFLYESFDYAEIKYINMNERNNVL--RKTSFSSYRNYYRYQFIQQLDRWGYRIISLG 233
K+ L E F + + YIN ERN++L + +FS RNYYRYQ IQQLD W +RIISLG
Sbjct: 181 KKNFLLNELFSFDVLYYIN--ERNSILLQQNINFSFSRNYYRYQLIQQLDFWSFRIISLG 238
Query: 234 FIFLTIGILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVA 293
FIFLT+GILSGAVWANE WGSYWNWDPKETWAFITWTIFAIYLH + N+ + G+NS+IVA
Sbjct: 239 FIFLTVGILSGAVWANETWGSYWNWDPKETWAFITWTIFAIYLHIKTNRNVRGINSAIVA 298
Query: 294 SIGFLIIWICYFGVNLLGIGLHNYGSFTSN 323
IGF++IWICYFGVNLLGIGLH+YGSFTSN
Sbjct: 299 LIGFILIWICYFGVNLLGIGLHSYGSFTSN 328
>UniRef100_Q8S8U8 CcsA protein [Atropa belladonna]
Length = 312
Score = 463 bits (1192), Expect = e-129
Identities = 233/322 (72%), Positives = 262/322 (81%), Gaps = 13/322 (4%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
MIFSTLEHILTHISFS+VSIVI+IHLIT V++IV YDSS+KG+I+TF CITGLL+TRW
Sbjct: 1 MIFSTLEHILTHISFSIVSIVITIHLITFLVDEIVKLYDSSEKGIIVTFFCITGLLVTRW 60
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAPSVIFTQGFATSGLLTE 120
SGH P SDLYESLIFLSW FS+ +++P FK LS I PS IFTQGFATSG+LTE
Sbjct: 61 ISSGHFPLSDLYESLIFLSWSFSLIHIIPYFKNNVLILSKITGPSAIFTQGFATSGILTE 120
Query: 121 MHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKSKRLSF 180
+HQS ILVPALQS WL+MHVSMMILGYAALLCGSLLSVA+LVITF++ + KS F
Sbjct: 121 IHQSGILVPALQSEWLIMHVSMMILGYAALLCGSLLSVALLVITFRKNRKLFYKSN--GF 178
Query: 181 LYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLTIG 240
+ ESF E NVL TSFS+ +NYYR Q IQQLD W YR+ISLGF FLTIG
Sbjct: 179 VNESFLLGE----------NVLENTSFSA-KNYYRSQLIQQLDYWSYRVISLGFTFLTIG 227
Query: 241 ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFLII 300
ILSGAVWANEAWGSYWNWDPKETWAFITW +FAIYLH+R N L G NS+I+A+IGFLII
Sbjct: 228 ILSGAVWANEAWGSYWNWDPKETWAFITWIVFAIYLHTRTNINLRGANSAIIATIGFLII 287
Query: 301 WICYFGVNLLGIGLHNYGSFTS 322
WICYFGVNLLGIGLH+YGSFTS
Sbjct: 288 WICYFGVNLLGIGLHSYGSFTS 309
>UniRef100_Q6EW01 CcsA protein [Nymphaea alba]
Length = 308
Score = 461 bits (1186), Expect = e-128
Identities = 226/323 (69%), Positives = 259/323 (79%), Gaps = 16/323 (4%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
MIF+TLEHILTHISFS++SIVI HL+TL V +IVG DSS+KGMI TF CITGLL+TRW
Sbjct: 1 MIFATLEHILTHISFSIISIVIPTHLMTL-VYEIVGLCDSSEKGMITTFFCITGLLVTRW 59
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAPSVIFTQGFATSGLLTE 120
+SGH+P SDLYESL+FLSW FS+ ++VP F+ KN S I APS I TQGFATSGLLT+
Sbjct: 60 IYSGHVPLSDLYESLMFLSWSFSLIHIVPYFRNYKNFFSKITAPSAILTQGFATSGLLTK 119
Query: 121 MHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKSKRLSF 180
MHQS ILVPALQS WLMMHVSMM+L YAALLCGSLLS+ +LVITF+ I I GK+ L
Sbjct: 120 MHQSAILVPALQSRWLMMHVSMMLLSYAALLCGSLLSITLLVITFRRKIDIFGKTNHL-- 177
Query: 181 LYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLTIG 240
L SF + E +Y+N S+RNY+RYQ Q+LD W YR+I LGF LTIG
Sbjct: 178 LISSFSFDETQYVNF-------------SFRNYHRYQLTQRLDYWSYRVIGLGFTLLTIG 224
Query: 241 ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFLII 300
ILSGAVWANEAWGSYWNWDPKETWAFITWT+FAIYLH+R NK L+G NS+IVAS+GFLII
Sbjct: 225 ILSGAVWANEAWGSYWNWDPKETWAFITWTVFAIYLHTRTNKSLQGANSAIVASMGFLII 284
Query: 301 WICYFGVNLLGIGLHNYGSFTSN 323
WICYFGVNLLG GLH+YGSFT N
Sbjct: 285 WICYFGVNLLGRGLHSYGSFTLN 307
>UniRef100_Q70XW4 CcsA protein [Amborella trichopoda]
Length = 313
Score = 457 bits (1175), Expect = e-127
Identities = 222/320 (69%), Positives = 263/320 (81%), Gaps = 10/320 (3%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
MIF+TLEHILTHISFS++SIVI+ HL+T F +I G D S+KGMI FL ITGLL+TRW
Sbjct: 1 MIFATLEHILTHISFSIISIVITTHLMT-FAREIAGLSDLSEKGMIAVFLRITGLLVTRW 59
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAPSVIFTQGFATSGLLTE 120
+SGHLP S+LYESLIFLSWGFS+ +M+ + + KN LS+I APS I TQGF TSGLLTE
Sbjct: 60 IYSGHLPLSNLYESLIFLSWGFSLIHMISKIQNHKNFLSSITAPSAILTQGFVTSGLLTE 119
Query: 121 MHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKSKRLSF 180
MHQS ILVPALQSHWL+MHVSMM+L YAALLCGSLLS+A+LVITF++ + I +
Sbjct: 120 MHQSAILVPALQSHWLIMHVSMMLLSYAALLCGSLLSIALLVITFRKKLDI------IFL 173
Query: 181 LYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLTIG 240
L F + EI+Y +NE+ ++L+ TSF S+RNY++YQ QQLD W YR+I +GF LT+G
Sbjct: 174 LIRLFSFGEIQY--LNEKRSILQNTSF-SFRNYHKYQLTQQLDHWSYRVIGIGFTLLTLG 230
Query: 241 ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFLII 300
ILSGAVWANEAWGSYWNWDPKETWAFITWTI AIYLH+R NK L+ NS+IVASIGFLII
Sbjct: 231 ILSGAVWANEAWGSYWNWDPKETWAFITWTISAIYLHTRTNKNLQSANSAIVASIGFLII 290
Query: 301 WICYFGVNLLGIGLHNYGSF 320
WICYFGVNLLGIGLH+YGSF
Sbjct: 291 WICYFGVNLLGIGLHSYGSF 310
>UniRef100_Q6ENA8 Heme attachment protein [Oryza nivara]
Length = 321
Score = 451 bits (1160), Expect = e-125
Identities = 217/321 (67%), Positives = 255/321 (78%), Gaps = 3/321 (0%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
M+F+TLEHILTHISFS +SIVI+IHLITL V ++ G DSS+KGMI TF CITG L++RW
Sbjct: 1 MLFATLEHILTHISFSTISIVITIHLITLLVRELGGLRDSSEKGMIATFFCITGFLVSRW 60
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAPSVIFTQGFATSGLLTE 120
SGH P S+LYESLIFLSW I +M+P+ + KNDLSTI PS I TQGFATSGLLTE
Sbjct: 61 ASSGHFPLSNLYESLIFLSWALYILHMIPKIQNSKNDLSTITTPSTILTQGFATSGLLTE 120
Query: 121 MHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKSKRLSF 180
MHQS ILVPALQS WLMMHVSMM+L YA LLCGSLLS A+L+I F++ + K K+ +
Sbjct: 121 MHQSTILVPALQSQWLMMHVSMMLLSYATLLCGSLLSAALLMIRFRKNLDFFSKKKK-NV 179
Query: 181 LYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLTIG 240
L ++F + EI+Y + + L+ T F + NYY+YQ I++LD W YR+ISLGF LTIG
Sbjct: 180 LLKTFFFNEIEY--FYAKRSALKSTFFPLFPNYYKYQLIERLDSWSYRVISLGFTLLTIG 237
Query: 241 ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFLII 300
IL GAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSR N +G S+ VASIGFLII
Sbjct: 238 ILCGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRTNPNWKGTKSAFVASIGFLII 297
Query: 301 WICYFGVNLLGIGLHNYGSFT 321
WICYFG+NLLGIGLH+YGSFT
Sbjct: 298 WICYFGINLLGIGLHSYGSFT 318
>UniRef100_P12215 Cytochrome c biogenesis protein ccsA [Oryza sativa]
Length = 321
Score = 451 bits (1159), Expect = e-125
Identities = 217/321 (67%), Positives = 255/321 (78%), Gaps = 3/321 (0%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
M+F+TLEHILTHISFS +SIVI+IHLITL V ++ G DSS+KGMI TF CITG L++RW
Sbjct: 1 MLFATLEHILTHISFSTISIVITIHLITLLVRELGGLRDSSEKGMIATFFCITGFLVSRW 60
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAPSVIFTQGFATSGLLTE 120
SGH P S+LYESLIFLSW I +M+P+ + KNDLSTI PS I TQGFATSGLLTE
Sbjct: 61 ASSGHFPLSNLYESLIFLSWALYILHMIPKIQNSKNDLSTITTPSTILTQGFATSGLLTE 120
Query: 121 MHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKSKRLSF 180
MHQS ILVPALQS WLMMHVSMM+L YA LLCGSLLS A+L+I F++ + K K+ +
Sbjct: 121 MHQSTILVPALQSQWLMMHVSMMLLSYATLLCGSLLSAALLMIRFRKNLDFFSKKKK-NV 179
Query: 181 LYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLTIG 240
L ++F + EI+Y + + L+ T F + NYY+YQ I++LD W YR+ISLGF LTIG
Sbjct: 180 LSKTFFFNEIEY--FYAKRSALKSTFFPLFPNYYKYQLIERLDSWSYRVISLGFTLLTIG 237
Query: 241 ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFLII 300
IL GAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSR N +G S+ VASIGFLII
Sbjct: 238 ILCGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRTNPNWKGTKSAFVASIGFLII 297
Query: 301 WICYFGVNLLGIGLHNYGSFT 321
WICYFG+NLLGIGLH+YGSFT
Sbjct: 298 WICYFGINLLGIGLHSYGSFT 318
>UniRef100_P46659 Cytochrome c biogenesis protein ccsA [Zea mays]
Length = 321
Score = 447 bits (1149), Expect = e-124
Identities = 214/321 (66%), Positives = 254/321 (78%), Gaps = 4/321 (1%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
M+F+TLEHILTHISFS +SIVI+IHLITL V ++ G DSS+KGMI TF ITG L++RW
Sbjct: 1 MLFATLEHILTHISFSTISIVITIHLITLLVRELRGLRDSSEKGMIATFFSITGFLVSRW 60
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAPSVIFTQGFATSGLLTE 120
SGH P S+LYESLIFLSW I + +P+ + KNDLSTI PS I TQGFATSGLLTE
Sbjct: 61 VSSGHFPLSNLYESLIFLSWTLYILHTIPKIQNSKNDLSTITTPSTILTQGFATSGLLTE 120
Query: 121 MHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKSKRLSF 180
MHQS ILVPALQS WLMMHVSMM+L YA LLCGSLLS A+L+I F++ K +
Sbjct: 121 MHQSTILVPALQSQWLMMHVSMMLLSYATLLCGSLLSAALLIIRFRKNFDFFSLKKNV-- 178
Query: 181 LYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLTIG 240
++F ++EI+Y + + + L+ TSF + NYY+YQ ++LD W YR+ISLGF LT+G
Sbjct: 179 FLKTFFFSEIEY--LYAKRSALKNTSFPVFPNYYKYQLTERLDSWSYRVISLGFTLLTVG 236
Query: 241 ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFLII 300
IL GAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKN +G NS++VASIGFLII
Sbjct: 237 ILCGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNPNWKGTNSALVASIGFLII 296
Query: 301 WICYFGVNLLGIGLHNYGSFT 321
WICYFG+NLLGIGLH+YGSFT
Sbjct: 297 WICYFGINLLGIGLHSYGSFT 317
>UniRef100_Q6ENP8 C-type cytochrome synthesis [Saccharum officinarum]
Length = 321
Score = 446 bits (1146), Expect = e-124
Identities = 214/321 (66%), Positives = 253/321 (78%), Gaps = 4/321 (1%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
M+F+TLEHILTHISFS +SIVI+IHLITL V ++ G DSS+KGMI TF ITG L++RW
Sbjct: 1 MLFATLEHILTHISFSTISIVITIHLITLLVRELRGLRDSSEKGMIATFFSITGFLVSRW 60
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAPSVIFTQGFATSGLLTE 120
SGH P S+LYESLIFLSW I + +P+ + KNDLSTI PS I TQGFATSGLLTE
Sbjct: 61 VSSGHFPLSNLYESLIFLSWTLYILHTIPKIQNSKNDLSTITTPSTILTQGFATSGLLTE 120
Query: 121 MHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKSKRLSF 180
MHQS ILVPALQS WLMMHVSMM+L YA LLCGSLLS A+L+I F+ K +
Sbjct: 121 MHQSTILVPALQSQWLMMHVSMMLLSYATLLCGSLLSAALLIIRFRNSFDFFSLKKNV-- 178
Query: 181 LYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLTIG 240
L ++F ++EI+Y + + + L+ TSF + NYY+YQ ++LD W YR+ISLGF LT+G
Sbjct: 179 LRKTFFFSEIEY--LYAKRSALKNTSFPVFPNYYKYQLTERLDSWSYRVISLGFTLLTVG 236
Query: 241 ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFLII 300
IL GAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSR N +G NS++VASIGFLII
Sbjct: 237 ILCGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRTNPNWKGTNSALVASIGFLII 296
Query: 301 WICYFGVNLLGIGLHNYGSFT 321
WICYFG+NLLGIGLH+YGSFT
Sbjct: 297 WICYFGINLLGIGLHSYGSFT 317
>UniRef100_P58266 Cytochrome c biogenesis protein ccsA [Triticum aestivum]
Length = 322
Score = 432 bits (1112), Expect = e-120
Identities = 209/321 (65%), Positives = 251/321 (78%), Gaps = 3/321 (0%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
M+F+TLEHILTHISFS +SIVI+IHLITL V ++ G DSS+KGMI+TF ITG L++RW
Sbjct: 1 MLFATLEHILTHISFSTISIVITIHLITLLVRELGGLRDSSEKGMIVTFFSITGFLVSRW 60
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAPSVIFTQGFATSGLLTE 120
SGH P S+LYESLI LSW I + +P+ + KNDLSTI PS I TQGFATSGLLTE
Sbjct: 61 ASSGHFPLSNLYESLISLSWALYILHTIPKIQNSKNDLSTITTPSTILTQGFATSGLLTE 120
Query: 121 MHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKSKRLSF 180
MHQS ILVPALQS WLMMHVSMM+L YA LL G LS AIL+I F+ K K+ +
Sbjct: 121 MHQSTILVPALQSQWLMMHVSMMLLSYATLLGGPPLSAAILIIRFRNNFHFFSKKKK-NV 179
Query: 181 LYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLTIG 240
L ++F +++IKY + + L++TS S+ NYY+YQ ++LD W YR+ISLGF LT G
Sbjct: 180 LNKTFLFSDIKY--FYAKRSALKRTSVPSFPNYYKYQLTERLDSWSYRVISLGFTLLTGG 237
Query: 241 ILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFLII 300
IL GAVWANEAWG+YWNWDPKETWAFITWTIFAIYLHSR + +G NS+++ASIGFLII
Sbjct: 238 ILGGAVWANEAWGAYWNWDPKETWAFITWTIFAIYLHSRTHPNWKGTNSALIASIGFLII 297
Query: 301 WICYFGVNLLGIGLHNYGSFT 321
WICYFG+NLLGIGLH+YGSFT
Sbjct: 298 WICYFGINLLGIGLHSYGSFT 318
>UniRef100_Q6QY38 Cytochrome c biogenesis protein [Oryza sativa]
Length = 285
Score = 400 bits (1027), Expect = e-110
Identities = 191/283 (67%), Positives = 223/283 (78%), Gaps = 3/283 (1%)
Query: 39 DSSKKGMIITFLCITGLLITRWFFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDL 98
DSS+KGMI TF CITG L++RW SGH P S+LYESLIFLSW I +M+P+ + KNDL
Sbjct: 3 DSSEKGMIATFFCITGFLVSRWASSGHFPLSNLYESLIFLSWALYILHMIPKIQNSKNDL 62
Query: 99 STIIAPSVIFTQGFATSGLLTEMHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSV 158
STI PS I TQGFATSGLLTEMHQS ILVPALQS WLMMHVSMM+L YA LLCGSLLS
Sbjct: 63 STITTPSTILTQGFATSGLLTEMHQSTILVPALQSQWLMMHVSMMLLSYATLLCGSLLSA 122
Query: 159 AILVITFQELIPILGKSKRLSFLYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQF 218
A+L+I F++ + K K+ + L ++F + EI+Y + + L+ T F + NYY+YQ
Sbjct: 123 ALLMIRFRKNLDFFSKKKK-NVLLKTFFFNEIEYFYA--KRSALKSTFFPLFPNYYKYQL 179
Query: 219 IQQLDRWGYRIISLGFIFLTIGILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHS 278
I++LD W YR+ISLGF LTIGIL GAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHS
Sbjct: 180 IERLDSWSYRVISLGFTLLTIGILCGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHS 239
Query: 279 RKNKKLEGLNSSIVASIGFLIIWICYFGVNLLGIGLHNYGSFT 321
R N +G S+ VASIGFLIIWICYFG+NLLGIGLH+YGSFT
Sbjct: 240 RTNPNWKGTKSAFVASIGFLIIWICYFGINLLGIGLHSYGSFT 282
>UniRef100_Q6QXR2 Cytochrome c biogenesis protein [Oryza sativa]
Length = 277
Score = 389 bits (1000), Expect = e-107
Identities = 186/277 (67%), Positives = 217/277 (78%), Gaps = 3/277 (1%)
Query: 45 MIITFLCITGLLITRWFFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAP 104
MI TF CITG L++RW SGH P S+LYESLIFLSW I +M+P+ + KNDLSTI P
Sbjct: 1 MIATFFCITGFLVSRWASSGHFPLSNLYESLIFLSWALYILHMIPKIQNSKNDLSTITTP 60
Query: 105 SVIFTQGFATSGLLTEMHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVIT 164
S I TQGFATSGLLTEMHQS ILVPALQS WLMMHVSMM+L YA LLCGSLLS A+L+I
Sbjct: 61 STILTQGFATSGLLTEMHQSTILVPALQSQWLMMHVSMMLLSYATLLCGSLLSAALLMIR 120
Query: 165 FQELIPILGKSKRLSFLYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDR 224
F++ + K K+ + L ++F + EI+Y + + L+ T F + NYY+YQ I++LD
Sbjct: 121 FRKNLDFFSKKKK-NVLSKTFFFNEIEYFYA--KRSALKSTFFPLFPNYYKYQLIERLDS 177
Query: 225 WGYRIISLGFIFLTIGILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKL 284
W YR+ISLGF LTIGIL GAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSR N
Sbjct: 178 WSYRVISLGFTLLTIGILCGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRTNPNW 237
Query: 285 EGLNSSIVASIGFLIIWICYFGVNLLGIGLHNYGSFT 321
+G S+ VASIGFLIIWICYFG+NLLGIGLH+YGSFT
Sbjct: 238 KGTKSAFVASIGFLIIWICYFGINLLGIGLHSYGSFT 274
>UniRef100_P41650 Cytochrome c biogenesis protein ccsA [Pinus thunbergii]
Length = 320
Score = 344 bits (883), Expect = 2e-93
Identities = 178/324 (54%), Positives = 224/324 (68%), Gaps = 14/324 (4%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
MIF TLEHIL HISFS++ +V I+ TL V +I G S KGMI+TFLC TGLLI RW
Sbjct: 1 MIFITLEHILAHISFSLILVVTLIYWGTL-VYRIEGLSSSGGKGMIVTFLCTTGLLINRW 59
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKND-LSTIIAPSVIFTQGFATSGLLT 119
+SGHLP S+LYES +FLSW S+F+++ + + + L I APS + T GFAT GL
Sbjct: 60 LYSGHLPLSNLYESFMFLSWSSSVFHILLEVRSRDDRWLGAITAPSAMLTHGFATLGLPE 119
Query: 120 EMHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQELIPILGKSKRLS 179
EM +S +LVPALQSHW MMHVSM++ YA LLCGSL S+A+LVI + G ++++
Sbjct: 120 EMQRSGMLVPALQSHWSMMHVSMILFSYATLLCGSLASIALLVI-------MSGVNRQVI 172
Query: 180 F-----LYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGF 234
F L+ + + ++ + L+ T + S NY + Q I+QLD W YR I LGF
Sbjct: 173 FGAMDNLFSRAILPNENFYSHEKQKSDLQYTVYFSSTNYRKCQLIKQLDHWSYRAIGLGF 232
Query: 235 IFLTIGILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVAS 294
TIG LSGA+WANEAWGSYW+WDPKETWA ITWTIFAIYLH+R NK +G +IVAS
Sbjct: 233 SLSTIGTLSGAIWANEAWGSYWSWDPKETWALITWTIFAIYLHTRMNKGWQGEEPAIVAS 292
Query: 295 IGFLIIWICYFGVNLLGIGLHNYG 318
+GF I+WI Y GVNLLGIGLH+YG
Sbjct: 293 LGFFIVWIRYLGVNLLGIGLHSYG 316
>UniRef100_P12214 Cytochrome c biogenesis protein ccsA [Marchantia polymorpha]
Length = 320
Score = 317 bits (812), Expect = 3e-85
Identities = 166/320 (51%), Positives = 210/320 (64%), Gaps = 5/320 (1%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
M F TLE IL H SF ++ V I+ I + M I IT L+ RW
Sbjct: 1 MPFITLERILAHTSFFLLFFVTFIYWGKFLYINIKPITILGEISMKIACFFITTFLLIRW 60
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRFKKQKNDLSTIIAPSVIFTQGFATSGLLTE 120
SGH P S+LYES +FLSW F++ +++ K + L I APS + T GFAT L E
Sbjct: 61 SSSGHFPLSNLYESSMFLSWSFTLIHLILENKSKNTWLGIITAPSAMLTHGFATLSLPKE 120
Query: 121 MHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITF--QELIPILGKSKRL 178
M +SV LVPALQSHWLMMHV+MM+L Y+ LLCGSLL++ IL+IT Q+ +PIL +
Sbjct: 121 MQESVFLVPALQSHWLMMHVTMMMLSYSTLLCGSLLAITILIITLTKQKNLPIL--TSYF 178
Query: 179 SFLYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISLGFIFLT 238
+F + SF + + NE + + FS + N+ ++Q I++LD W YR+ISLGF LT
Sbjct: 179 NFPFNSFIFKNLLQPMENEILSYKTQKVFS-FINFRKWQLIKELDNWSYRVISLGFPLLT 237
Query: 239 IGILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIVASIGFL 298
IGILSGAVWANEAWGSYWNWDPKETWA ITW IFAIYLH+R K +G +I+AS+GF
Sbjct: 238 IGILSGAVWANEAWGSYWNWDPKETWALITWLIFAIYLHTRMIKGWQGKKPAIIASLGFF 297
Query: 299 IIWICYFGVNLLGIGLHNYG 318
I+WICY GVNLLG GLH+YG
Sbjct: 298 IVWICYLGVNLLGKGLHSYG 317
>UniRef100_Q5SCZ5 Cytochrome c heme attachment protein [Huperzia lucidula]
Length = 327
Score = 310 bits (794), Expect = 3e-83
Identities = 166/326 (50%), Positives = 207/326 (62%), Gaps = 14/326 (4%)
Query: 1 MIFSTLEHILTHISFSVVSIVISIHLITLFVNQIVGFYDSSKKGMIITFLCITGLLITRW 60
MI TLEHIL H F ++ ++ TL + + MII F+CITG +TRW
Sbjct: 1 MILVTLEHILAHTPFFLLFFATLLYWGTLVFAKNNKLSSLGQISMIIAFICITGFSLTRW 60
Query: 61 FFSGHLPFSDLYESLIFLSWGFSIFYMVPRF------KKQKNDLSTIIAPSVIFTQGFAT 114
+SGHLPFS+LYES +FLSW + +++ K + N L TI PS + T GFAT
Sbjct: 61 SYSGHLPFSNLYESFMFLSWSLCLIHIIVENSIIVGNKSKINWLGTITVPSAMLTHGFAT 120
Query: 115 SGLLTEMHQSVILVPALQSHWLMMHVSMMILGYAALLCGSLLSVAILVITFQEL--IPIL 172
GL EM QS +LVPALQSHWLMMHVSMM+L YA LCGS L++A VI ++ IP+L
Sbjct: 121 LGLPREMQQSTVLVPALQSHWLMMHVSMMMLSYATPLCGSSLAIAFPVIASKKHVEIPVL 180
Query: 173 GKSKRLSFLYESFDYAEIKYINMNERNNVLRKTSFSSYRNYYRYQFIQQLDRWGYRIISL 232
G + + F + S + N ++ + L TS NY + Q QQLD W YRIISL
Sbjct: 181 GANTKPYFWFFSLEE------NDKQKESFLSNTSAPLSINYNKSQLTQQLDHWSYRIISL 234
Query: 233 GFIFLTIGILSGAVWANEAWGSYWNWDPKETWAFITWTIFAIYLHSRKNKKLEGLNSSIV 292
GF TIGILSGAVWANEAWGS+WNWDPKETWA +TW FAI+LH+R K +G +IV
Sbjct: 235 GFPLSTIGILSGAVWANEAWGSHWNWDPKETWALVTWLAFAIHLHTRITKGWQGKKPAIV 294
Query: 293 ASIGFLIIWICYFGVNLLGIGLHNYG 318
AS+GF II IC+ GVN G GLH+YG
Sbjct: 295 ASLGFSIIRICHLGVNPSGKGLHSYG 320
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.329 0.142 0.448
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 532,731,182
Number of Sequences: 2790947
Number of extensions: 22133666
Number of successful extensions: 87129
Number of sequences better than 10.0: 1068
Number of HSP's better than 10.0 without gapping: 909
Number of HSP's successfully gapped in prelim test: 159
Number of HSP's that attempted gapping in prelim test: 85306
Number of HSP's gapped (non-prelim): 1377
length of query: 323
length of database: 848,049,833
effective HSP length: 127
effective length of query: 196
effective length of database: 493,599,564
effective search space: 96745514544
effective search space used: 96745514544
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 75 (33.5 bits)
Description of BAB33244.1

