
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
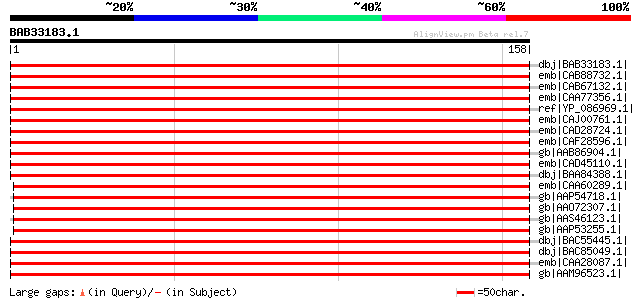
Query= BAB33183.1 158 aa
(158 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB33183.1| NADH dehydrogenase subunit [Lotus corniculatus v... 340 6e-93
emb|CAB88732.1| NADH dehydrogenase 19kDa subunit [Spinacia olera... 320 6e-87
emb|CAB67132.1| NADH-plastoquinone oxidoreductase subunit J [Oen... 320 8e-87
emb|CAA77356.1| NADH dehydrogenase 30kD subunit [Nicotiana tabac... 317 5e-86
ref|YP_086969.1| NADH dehydrogenase 19 kDa subunit [Panax ginsen... 314 4e-85
emb|CAJ00761.1| NADH dehydrogenase subunit J [Cucumis sativus] 311 5e-84
emb|CAD28724.1| NADH dehydrogenase 19kDa subunit [Calycanthus fl... 311 5e-84
emb|CAF28596.1| NADH dehydrogenase 19kDa subunit [Nymphaea alba]... 310 6e-84
gb|AAB86904.1| NADH plastiquinone oxidoreductase subunit J [Lupi... 308 3e-83
emb|CAD45110.1| NADH dehydrogenase 19kDa subunit [Amborella tric... 305 2e-82
dbj|BAA84388.1| NADH dehydrogenase subunit [Arabidopsis thaliana... 303 1e-81
emb|CAA60289.1| ndhJ [Zea mays] gi|48478676|gb|AAT44696.1| NADH ... 291 5e-78
gb|AAP54718.1| NADH-plastoquinone oxidoreductase subunit J [Oryz... 290 7e-78
gb|AAO72307.1| NdhJ [Hordeum vulgare] gi|14017575|ref|NP_114262.... 290 7e-78
gb|AAS46123.1| NADH dehydrogenase subunit J [Oryza sativa (japon... 290 7e-78
gb|AAP53255.1| hypothetical protein ORF159 from chromosome 10 ch... 288 4e-77
dbj|BAC55445.1| NADH dehydrogenase 30 kDa subunit [Anthoceros fo... 282 2e-75
dbj|BAC85049.1| NADH dehydrogenase 30 kD subunit [Physcomitrella... 276 1e-73
emb|CAA28087.1| unnamed protein product [Marchantia polymorpha] ... 275 3e-73
gb|AAM96523.1| subunit J of NADH-plastoquinone oxidoreductase [C... 269 2e-71
>dbj|BAB33183.1| NADH dehydrogenase subunit [Lotus corniculatus var. japonicus]
gi|13518425|ref|NP_084785.1| NADH dehydrogenase subunit
J [Lotus corniculatus var. japonicus]
gi|23821910|sp|Q9BBT6|NUGC_LOTJA NAD(P)H-quinone
oxidoreductase chain J, chloroplast (NAD(P)H
dehydrogenase, chain J) (NADH-plastoquinone
oxidoreductase subunit J)
Length = 158
Score = 340 bits (873), Expect = 6e-93
Identities = 158/158 (100%), Positives = 158/158 (100%)
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS
Sbjct: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI
Sbjct: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
Query: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH
Sbjct: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
>emb|CAB88732.1| NADH dehydrogenase 19kDa subunit [Spinacia oleracea]
gi|11497531|ref|NP_054939.1| NADH dehydrogenase subunit
J [Spinacia oleracea] gi|18203266|sp|Q9M3M1|NUGC_SPIOL
NAD(P)H-quinone oxidoreductase chain J, chloroplast
(NAD(P)H dehydrogenase, chain J) (NADH-plastoquinone
oxidoreductase subunit J)
Length = 158
Score = 320 bits (821), Expect = 6e-87
Identities = 147/158 (93%), Positives = 152/158 (96%)
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDV+
Sbjct: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVA 60
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
PGGLLASVYHLTRIE G+ QPEEVCIK+F PR NPRIPS+FWVWKSADFQERESYDM GI
Sbjct: 61 PGGLLASVYHLTRIEYGVDQPEEVCIKVFAPRRNPRIPSVFWVWKSADFQERESYDMFGI 120
Query: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
SYDNHPRLKRILMPESWIGWPLRKDYI PNFYEIQDA+
Sbjct: 121 SYDNHPRLKRILMPESWIGWPLRKDYIVPNFYEIQDAY 158
>emb|CAB67132.1| NADH-plastoquinone oxidoreductase subunit J [Oenothera elata subsp.
hookeri] gi|13518308|ref|NP_084667.1| NADH dehydrogenase
subunit J [Oenothera elata subsp. hookeri]
gi|22256941|sp|Q9MTP3|NUGC_OENHO NAD(P)H-quinone
oxidoreductase chain J, chloroplast (NAD(P)H
dehydrogenase, chain J) (NADH-plastoquinone
oxidoreductase subunit J)
Length = 158
Score = 320 bits (820), Expect = 8e-87
Identities = 147/158 (93%), Positives = 152/158 (96%)
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPE+WHSIAVILYVYGYNYLRSQCAYDV+
Sbjct: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEEWHSIAVILYVYGYNYLRSQCAYDVA 60
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
PGGLLASVYHLTRIE G+ Q EEVCIK+F PRNNPRIPS+FWVWKSADFQERESYDMLGI
Sbjct: 61 PGGLLASVYHLTRIEYGVDQAEEVCIKVFAPRNNPRIPSVFWVWKSADFQERESYDMLGI 120
Query: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
YDNHPRLKRILMPESWIGWPLRKDYI PNFYEIQDAH
Sbjct: 121 RYDNHPRLKRILMPESWIGWPLRKDYIAPNFYEIQDAH 158
>emb|CAA77356.1| NADH dehydrogenase 30kD subunit [Nicotiana tabacum]
gi|20068334|emb|CAC88047.1| NADH dehydrogenase 19kD
subunit [Atropa belladonna] gi|28261720|ref|NP_783235.1|
NADH dehydrogenase subunit J [Atropa belladonna]
gi|11465960|ref|NP_054502.1| NADH dehydrogenase subunit
J [Nicotiana tabacum] gi|128859|sp|P12201|NUGC_TOBAC
NAD(P)H-quinone oxidoreductase chain J, chloroplast
(NAD(P)H dehydrogenase, chain J) (NADH-plastoquinone
oxidoreductase subunit J) gi|82206|pir||A05194
hypothetical protein 158 - common tobacco chloroplast
gi|225204|prf||1211235AJ ORF 158
Length = 158
Score = 317 bits (813), Expect = 5e-86
Identities = 145/158 (91%), Positives = 150/158 (94%)
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
MQGRLSAWLVKHGL+HRSLGFDYQGIETLQIKPEDWHSIAVI YVYGYNYLRSQCAYDV+
Sbjct: 1 MQGRLSAWLVKHGLIHRSLGFDYQGIETLQIKPEDWHSIAVIFYVYGYNYLRSQCAYDVA 60
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
PGGLLASVYHLTRIE G+ QPEEVCIK+F R NPRIPS+FWVWKS DFQERESYDMLGI
Sbjct: 61 PGGLLASVYHLTRIEDGVDQPEEVCIKVFASRRNPRIPSVFWVWKSVDFQERESYDMLGI 120
Query: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
SYDNHPRLKRILMPESWIGWPLRKDYI PNFYEIQDAH
Sbjct: 121 SYDNHPRLKRILMPESWIGWPLRKDYIAPNFYEIQDAH 158
>ref|YP_086969.1| NADH dehydrogenase 19 kDa subunit [Panax ginseng]
gi|51235316|gb|AAT98512.1| NADH dehydrogenase 19 kDa
subunit [Panax ginseng]
Length = 158
Score = 314 bits (805), Expect = 4e-85
Identities = 143/158 (90%), Positives = 149/158 (93%)
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
MQGRLSAWLVKHGL+HRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDV+
Sbjct: 1 MQGRLSAWLVKHGLIHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVA 60
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
PGGLLASVYHLTRIE G+ QPEEVCIK+F PR NPRIPS+FWVWKS DFQERESYDMLGI
Sbjct: 61 PGGLLASVYHLTRIEYGVDQPEEVCIKVFAPRRNPRIPSVFWVWKSVDFQERESYDMLGI 120
Query: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
Y HPRLKRILMPESW+GWPLRKDYI PNFYEIQDAH
Sbjct: 121 FYATHPRLKRILMPESWVGWPLRKDYIAPNFYEIQDAH 158
>emb|CAJ00761.1| NADH dehydrogenase subunit J [Cucumis sativus]
Length = 158
Score = 311 bits (796), Expect = 5e-84
Identities = 143/158 (90%), Positives = 149/158 (93%)
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
MQG LS+WLVKHGLVHRSLGFDYQGIETLQIKPE+WHSIAVILYVYGYNYLRSQCAYDV+
Sbjct: 1 MQGNLSSWLVKHGLVHRSLGFDYQGIETLQIKPEEWHSIAVILYVYGYNYLRSQCAYDVA 60
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
PGGLLASVYHLTRIE GI QPEEVCIK+F R NPRIPS+FWVWKSADF ERESYDMLGI
Sbjct: 61 PGGLLASVYHLTRIEYGIDQPEEVCIKVFAARINPRIPSVFWVWKSADFPERESYDMLGI 120
Query: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
YDNHPRLKRILMPESW+GWPLRKDYI PNFYEIQDAH
Sbjct: 121 YYDNHPRLKRILMPESWVGWPLRKDYIAPNFYEIQDAH 158
>emb|CAD28724.1| NADH dehydrogenase 19kDa subunit [Calycanthus floridus var.
glaucus] gi|32480846|ref|NP_862757.1| NADH dehydrogenase
subunit J [Calycanthus floridus var. glaucus]
Length = 158
Score = 311 bits (796), Expect = 5e-84
Identities = 141/158 (89%), Positives = 151/158 (95%)
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDW+SIAVI YVYGYNYLR QCAYDV+
Sbjct: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWYSIAVISYVYGYNYLRFQCAYDVA 60
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
PGGLLASVY+LTRI+ G+ QPEEVCIK+F PR NP+IPS+FW+WKSADFQERESYDMLGI
Sbjct: 61 PGGLLASVYYLTRIQYGVDQPEEVCIKVFAPRRNPKIPSVFWIWKSADFQERESYDMLGI 120
Query: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
SY+NHPRLKRILMPESWIGWPLRKDYI PNFYEIQDAH
Sbjct: 121 SYENHPRLKRILMPESWIGWPLRKDYIAPNFYEIQDAH 158
>emb|CAF28596.1| NADH dehydrogenase 19kDa subunit [Nymphaea alba]
gi|50346785|ref|YP_053158.1| NADH dehydrogenase 19kDa
subunit [Nymphaea alba]
Length = 158
Score = 310 bits (795), Expect = 6e-84
Identities = 140/158 (88%), Positives = 152/158 (95%)
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
MQGR SAWLVKH LVHRSLGFDYQG+ETLQIKPEDW+SIAVI YVYGYNYLRSQCAYDV+
Sbjct: 1 MQGRSSAWLVKHELVHRSLGFDYQGVETLQIKPEDWYSIAVISYVYGYNYLRSQCAYDVA 60
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
PGGLLASVYHLTRI+ G+ QPEEVCIK+FVPR+NPRIPS+FW+WKSADFQERESYDMLGI
Sbjct: 61 PGGLLASVYHLTRIQYGVDQPEEVCIKVFVPRSNPRIPSVFWIWKSADFQERESYDMLGI 120
Query: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
SYDNHPR+KRILMPESWIGWPLRKDYI PNFYE+QDA+
Sbjct: 121 SYDNHPRMKRILMPESWIGWPLRKDYIAPNFYELQDAY 158
>gb|AAB86904.1| NADH plastiquinone oxidoreductase subunit J [Lupinus luteus]
gi|7459649|pir||T09639 NADH2 dehydrogenase (ubiquinone)
(EC 1.6.5.3) chain J - yellow lupine chloroplast
gi|3914184|sp|P92309|NUGC_LUPLU NAD(P)H-quinone
oxidoreductase chain J, chloroplast (NAD(P)H
dehydrogenase, chain J) (NADH-plastoquinone
oxidoreductase subunit J)
Length = 158
Score = 308 bits (789), Expect = 3e-83
Identities = 145/158 (91%), Positives = 148/158 (92%)
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
MQGRLSAWLVKHGL+HRSLGFDYQGIETLQIKPE WHSIAVILYVYGYNYLRSQCAYDV+
Sbjct: 1 MQGRLSAWLVKHGLIHRSLGFDYQGIETLQIKPEAWHSIAVILYVYGYNYLRSQCAYDVA 60
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
PGGLLASVYHLTRIECGI QPEEVCIK+FV IPSIFWVWKSADFQERESYDMLGI
Sbjct: 61 PGGLLASVYHLTRIECGIDQPEEVCIKVFVQGKILGIPSIFWVWKSADFQERESYDMLGI 120
Query: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
SY NHPRLKRILMPESWIGWPLRKDYI PNFYEIQDAH
Sbjct: 121 SYYNHPRLKRILMPESWIGWPLRKDYIAPNFYEIQDAH 158
>emb|CAD45110.1| NADH dehydrogenase 19kDa subunit [Amborella trichopoda]
gi|34500917|ref|NP_904102.1| NADH dehydrogenase 19kDa
subunit [Amborella trichopoda]
Length = 158
Score = 305 bits (782), Expect = 2e-82
Identities = 140/158 (88%), Positives = 148/158 (93%)
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
MQGRLSAWLVKH LVHRSLGFDYQGIETLQIK EDW+SIAVI YVYGYNYLRSQCAYDV+
Sbjct: 1 MQGRLSAWLVKHELVHRSLGFDYQGIETLQIKSEDWYSIAVISYVYGYNYLRSQCAYDVA 60
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
PGGLLASVYHLTRI+ G+ QPEEVCIK+F R NPRIPS+FW+WKS+DFQERESYDMLGI
Sbjct: 61 PGGLLASVYHLTRIQYGVDQPEEVCIKVFAQRRNPRIPSVFWIWKSSDFQERESYDMLGI 120
Query: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
YDNHPRLKRILMPESWIGWPLRKDYI PNFYEIQDAH
Sbjct: 121 YYDNHPRLKRILMPESWIGWPLRKDYIAPNFYEIQDAH 158
>dbj|BAA84388.1| NADH dehydrogenase subunit [Arabidopsis thaliana]
gi|7525036|ref|NP_051062.1| NADH dehydrogenase subunit J
[Arabidopsis thaliana] gi|6685680|sp|P56754|NUGC_ARATH
NAD(P)H-quinone oxidoreductase chain J, chloroplast
(NAD(P)H dehydrogenase, chain J) (NADH-plastoquinone
oxidoreductase subunit J)
Length = 158
Score = 303 bits (776), Expect = 1e-81
Identities = 139/158 (87%), Positives = 147/158 (92%)
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
MQG LS WL K GLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDV+
Sbjct: 1 MQGTLSVWLAKRGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVA 60
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
PGGLLASVYHLTRIE G+ Q EEVCIK+F R+NPRIPS+FWVWKS DFQERESYDMLGI
Sbjct: 61 PGGLLASVYHLTRIEYGVNQAEEVCIKVFTHRSNPRIPSVFWVWKSTDFQERESYDMLGI 120
Query: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
+YD+HPRLKRILMPESWIGWPLRKDYI PNFYEIQDA+
Sbjct: 121 TYDSHPRLKRILMPESWIGWPLRKDYIAPNFYEIQDAY 158
>emb|CAA60289.1| ndhJ [Zea mays] gi|48478676|gb|AAT44696.1| NADH dehydrogenase
subunit J [Saccharum hybrid cultivar SP-80-3280]
gi|50812531|ref|YP_054634.1| NADH dehydrogenase 30kD
subunit [Saccharum officinarum]
gi|11467195|ref|NP_043028.1| NADH dehydrogenase subunit
J [Zea mays] gi|48478774|ref|YP_024382.1| NADH
dehydrogenase subunit J [Saccharum hybrid cultivar
SP-80-3280] gi|12428|emb|CAA35483.1| unnamed protein
product [Zea mays] gi|1363530|pir||S58555 ndhJ protein -
maize chloroplast gi|49659515|dbj|BAD27296.1| NADH
dehydrogenase 30kD subunit [Saccharum officinarum]
gi|128855|sp|P19124|NUGC_MAIZE NAD(P)H-quinone
oxidoreductase chain J, chloroplast (NAD(P)H
dehydrogenase, chain J) (NADH-plastoquinone
oxidoreductase subunit J)
Length = 159
Score = 291 bits (744), Expect = 5e-78
Identities = 133/157 (84%), Positives = 145/157 (91%)
Query: 2 QGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVSP 61
QG LS WLVKH +VHRSLGFD++G+ETLQIK DW SIAVILYVYGYNYLRSQCAYDV+P
Sbjct: 3 QGWLSNWLVKHDVVHRSLGFDHRGVETLQIKAGDWDSIAVILYVYGYNYLRSQCAYDVAP 62
Query: 62 GGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGIS 121
GG LASVYHLTRI+ GI PEEVCIK+F ++NPRIPS+FWVW+SADFQERESYDM+GIS
Sbjct: 63 GGSLASVYHLTRIQYGIDNPEEVCIKVFAQKDNPRIPSVFWVWRSADFQERESYDMVGIS 122
Query: 122 YDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
YDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH
Sbjct: 123 YDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 159
>gb|AAP54718.1| NADH-plastoquinone oxidoreductase subunit J [Oryza sativa (japonica
cultivar-group)] gi|11987|emb|CAA33999.1| unnamed
protein product [Oryza sativa (japonica cultivar-group)]
gi|37536258|ref|NP_922431.1| NADH-plastoquinone
oxidoreductase subunit J [Oryza sativa (japonica
cultivar-group)] gi|20146756|gb|AAM12492.1|
NADH-plastoquinone oxidoreductase subunit J [Oryza
sativa (japonica cultivar-group)]
gi|56784293|dbj|BAD81975.1| Chloroplast NADH
dehydrogenase subunit J [Oryza sativa (japonica
cultivar-group)] gi|11466790|ref|NP_039386.1| NADH
dehydrogenase subunit J [Oryza sativa (japonica
cultivar-group)] gi|50233974|ref|YP_052752.1| NADH
dehydrogenase subunit J [Oryza nivara]
gi|49614998|dbj|BAD26781.1| NADH dehydrogenase subunit J
[Oryza nivara] gi|82539|pir||S05106 hypothetical protein
159 - rice chloroplast gi|128857|sp|P12200|NUGC_ORYSA
NAD(P)H-quinone oxidoreductase chain J, chloroplast
(NAD(P)H dehydrogenase, chain J) (NADH-plastoquinone
oxidoreductase subunit J) gi|226608|prf||1603356AE ORF
159
Length = 159
Score = 290 bits (743), Expect = 7e-78
Identities = 132/157 (84%), Positives = 146/157 (92%)
Query: 2 QGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVSP 61
QG LS WLVKH +VHRSLGFD++GIETLQIK EDW SIAVILYVYGYNYLRSQCAYDV+P
Sbjct: 3 QGWLSNWLVKHEVVHRSLGFDHRGIETLQIKAEDWDSIAVILYVYGYNYLRSQCAYDVAP 62
Query: 62 GGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGIS 121
GG LASVYHLTRI+ GI PEEVCIK+F ++NPRIPS+FW+W+S+DFQERES+DM+GIS
Sbjct: 63 GGSLASVYHLTRIQYGIDNPEEVCIKVFAQKDNPRIPSVFWIWRSSDFQERESFDMVGIS 122
Query: 122 YDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
YDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH
Sbjct: 123 YDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 159
>gb|AAO72307.1| NdhJ [Hordeum vulgare] gi|14017575|ref|NP_114262.1| NADH
dehydrogenase subunit J [Triticum aestivum]
gi|20178002|sp|Q95H60|NUGC_WHEAT NAD(P)H-quinone
oxidoreductase chain J, chloroplast (NAD(P)H
dehydrogenase, chain J) (NADH-plastoquinone
oxidoreductase subunit J) gi|13928208|dbj|BAB47037.1|
NADH dehydrogenase 30kDa subunit [Triticum aestivum]
Length = 159
Score = 290 bits (743), Expect = 7e-78
Identities = 133/157 (84%), Positives = 145/157 (91%)
Query: 2 QGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVSP 61
QG LS WLVKH +VHRSLGFD++GIETLQIK DW SIAVILYVYGYNYLRSQCAYDV+P
Sbjct: 3 QGWLSNWLVKHEVVHRSLGFDHRGIETLQIKAGDWDSIAVILYVYGYNYLRSQCAYDVAP 62
Query: 62 GGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGIS 121
GG LASVYHLTRI+ GI PEEVCIK+F ++NPRIPS+FW+W+SADFQERESYDM+GIS
Sbjct: 63 GGSLASVYHLTRIQYGIDNPEEVCIKVFAQKDNPRIPSVFWIWRSADFQERESYDMVGIS 122
Query: 122 YDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
YDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH
Sbjct: 123 YDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 159
>gb|AAS46123.1| NADH dehydrogenase subunit J [Oryza sativa (japonica
cultivar-group)] gi|42795621|gb|AAS46186.1| NADH
dehydrogenase subunit J [Oryza sativa (japonica
cultivar-group)] gi|42795490|gb|AAS46057.1| NADH
dehydrogenase subunit J [Oryza sativa (indica
cultivar-group)]
Length = 188
Score = 290 bits (743), Expect = 7e-78
Identities = 132/157 (84%), Positives = 146/157 (92%)
Query: 2 QGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVSP 61
QG LS WLVKH +VHRSLGFD++GIETLQIK EDW SIAVILYVYGYNYLRSQCAYDV+P
Sbjct: 32 QGWLSNWLVKHEVVHRSLGFDHRGIETLQIKAEDWDSIAVILYVYGYNYLRSQCAYDVAP 91
Query: 62 GGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGIS 121
GG LASVYHLTRI+ GI PEEVCIK+F ++NPRIPS+FW+W+S+DFQERES+DM+GIS
Sbjct: 92 GGSLASVYHLTRIQYGIDNPEEVCIKVFAQKDNPRIPSVFWIWRSSDFQERESFDMVGIS 151
Query: 122 YDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
YDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH
Sbjct: 152 YDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 188
>gb|AAP53255.1| hypothetical protein ORF159 from chromosome 10 chloroplast
insertion [Oryza sativa (japonica cultivar-group)]
gi|37533332|ref|NP_920968.1| hypothetical protein ORF159
from chromosome 10 chloroplast insertion [Oryza sativa
(japonica cultivar-group)] gi|21327361|gb|AAM48266.1|
Hypothetical protein ORF159 from chromosome 10
chloroplast insertion [Oryza sativa (japonica
cultivar-group)] gi|19920169|gb|AAM08601.1| Hypothetical
protein ORF159 from chromosome 10 chloroplast insertion
[Oryza sativa (japonica cultivar-group)]
Length = 159
Score = 288 bits (736), Expect = 4e-77
Identities = 131/157 (83%), Positives = 145/157 (91%)
Query: 2 QGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVSP 61
QG LS WLVKH +VHRSLGFD++GI TLQIK EDW SIAVILYVYGYNYLRSQCAYDV+P
Sbjct: 3 QGWLSNWLVKHEVVHRSLGFDHRGIGTLQIKAEDWDSIAVILYVYGYNYLRSQCAYDVAP 62
Query: 62 GGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGIS 121
GG LASVYHLTRI+ GI PEEVCIK+F ++NPRIPS+FW+W+S+DFQERES+DM+GIS
Sbjct: 63 GGSLASVYHLTRIQYGIDNPEEVCIKVFAQKDNPRIPSVFWIWRSSDFQERESFDMVGIS 122
Query: 122 YDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
YDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH
Sbjct: 123 YDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 159
>dbj|BAC55445.1| NADH dehydrogenase 30 kDa subunit [Anthoceros formosae]
gi|28202175|ref|NP_777416.1| NADH dehydrogenase subunit
J [Anthoceros formosae] gi|27807877|dbj|BAC55352.1| NADH
dehydrogenase 30 kDa subunit [Anthoceros formosae]
gi|46577626|sp|Q85BB2|NUGC_ANTFO NAD(P)H-quinone
oxidoreductase chain J, chloroplast (NAD(P)H
dehydrogenase, chain J) (NADH-plastoquinone
oxidoreductase subunit J)
Length = 169
Score = 282 bits (722), Expect = 2e-75
Identities = 125/158 (79%), Positives = 142/158 (89%)
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
+QGRLSAWL+KH L HR LGFDYQG+ETLQ++ EDW SIAV LY YG+NYLRSQC YDV+
Sbjct: 12 IQGRLSAWLIKHRLAHRPLGFDYQGVETLQVRSEDWLSIAVALYAYGFNYLRSQCVYDVA 71
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
PGGLLASVYHLT+++ QPEEVCIKIFV R NP+IPS+FWVWK ADFQERESYDMLGI
Sbjct: 72 PGGLLASVYHLTKVQSNADQPEEVCIKIFVSRKNPKIPSVFWVWKGADFQERESYDMLGI 131
Query: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
SY++HPRLKRILMP+SWIGWPLRKDYI PNFYE+QDA+
Sbjct: 132 SYESHPRLKRILMPDSWIGWPLRKDYIVPNFYELQDAY 169
>dbj|BAC85049.1| NADH dehydrogenase 30 kD subunit [Physcomitrella patens subsp.
patens] gi|34501412|ref|NP_904199.1| NADH dehydrogenase
subunit J [Physcomitrella patens subsp. patens]
Length = 169
Score = 276 bits (706), Expect = 1e-73
Identities = 123/158 (77%), Positives = 140/158 (87%)
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
+QGRLS WL H L HR LGFDYQG+E LQI+ EDW SIAV LYVYG+NYLRSQCAYDV+
Sbjct: 12 IQGRLSVWLANHKLPHRPLGFDYQGVEILQIRSEDWLSIAVALYVYGFNYLRSQCAYDVA 71
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
PGGLLASVYH T+IE + QPEE+CIKIFV R P+IPS+FW+WKSADFQERESYDMLGI
Sbjct: 72 PGGLLASVYHFTKIEDNVDQPEEICIKIFVSRQKPKIPSVFWIWKSADFQERESYDMLGI 131
Query: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
SY+NHPRLKRILMP++WIGWPLRKDYI P+FYE+QDA+
Sbjct: 132 SYENHPRLKRILMPDTWIGWPLRKDYIVPDFYELQDAY 169
>emb|CAA28087.1| unnamed protein product [Marchantia polymorpha]
gi|81336|pir||A05042 hypothetical protein 169 -
liverwort (Marchantia polymorpha) chloroplast
gi|11466705|ref|NP_039301.1| NADH dehydrogenase subunit
J [Marchantia polymorpha] gi|128856|sp|P12199|NUGC_MARPO
NAD(P)H-quinone oxidoreductase chain J, chloroplast
(NAD(P)H dehydrogenase, chain J) (NADH-plastoquinone
oxidoreductase subunit J)
Length = 169
Score = 275 bits (703), Expect = 3e-73
Identities = 124/158 (78%), Positives = 138/158 (86%)
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
+QGRLS WL+KH L HR LGFDYQGIETLQI+ EDW S+AV LYVYG+NYLRSQCAYDV
Sbjct: 12 IQGRLSIWLIKHNLKHRPLGFDYQGIETLQIRSEDWPSLAVALYVYGFNYLRSQCAYDVE 71
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
PGGLLASVYH T+I QPEE+CIKIF+ R NP+IPSIFWVWKSADFQERESYDM GI
Sbjct: 72 PGGLLASVYHFTKITDNADQPEEICIKIFILRKNPKIPSIFWVWKSADFQERESYDMFGI 131
Query: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
Y+NHP LKRILMP+SW+GWPLRKDYI PNFYE+QDA+
Sbjct: 132 FYENHPCLKRILMPDSWLGWPLRKDYIVPNFYELQDAY 169
>gb|AAM96523.1| subunit J of NADH-plastoquinone oxidoreductase [Chaetosphaeridium
globosum] gi|22711919|ref|NP_683806.1| NADH
dehydrogenase subunit J [Chaetosphaeridium globosum]
Length = 168
Score = 269 bits (688), Expect = 2e-71
Identities = 120/158 (75%), Positives = 140/158 (87%)
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
+QGR+SAWL + L HR LGFDYQG+ETLQIKPEDW SIAV LYV G+NYLR QC YDV
Sbjct: 11 VQGRISAWLASNNLPHRPLGFDYQGVETLQIKPEDWPSIAVALYVNGFNYLRCQCGYDVY 70
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
PGG LASVY+LT+++ + QPEEVCIKIFVPR NP+IPSIFWVWK+AD+QERE+YDMLGI
Sbjct: 71 PGGPLASVYYLTKVDDNVDQPEEVCIKIFVPRENPKIPSIFWVWKTADYQERETYDMLGI 130
Query: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
Y++HP LKRILMPESW+GWPLRKDYITP+FYE+QDA+
Sbjct: 131 VYESHPNLKRILMPESWLGWPLRKDYITPDFYELQDAY 168
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.324 0.143 0.474
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 319,103,590
Number of Sequences: 2540612
Number of extensions: 13410369
Number of successful extensions: 19499
Number of sequences better than 10.0: 359
Number of HSP's better than 10.0 without gapping: 343
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 19138
Number of HSP's gapped (non-prelim): 366
length of query: 158
length of database: 863,360,394
effective HSP length: 117
effective length of query: 41
effective length of database: 566,108,790
effective search space: 23210460390
effective search space used: 23210460390
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 69 (31.2 bits)
Description of BAB33183.1

