
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BAB33240.1 155 aa
(155 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
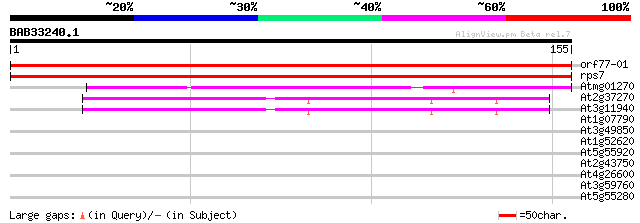
Score E
Sequences producing significant alignments: (bits) Value
orf77-01 -chloroplast genome- ribosomal protein S7 300 2e-82
rps7 -chloroplast genome- ribosomal protein S7 300 2e-82
Atmg01270 -mitochondrial genome- ribosomal protein S7 (RPS7) 76 6e-15
At2g37270 40S ribosomal protein S5 60 5e-10
At3g11940 putative 40S ribosomal protein S5 59 8e-10
At1g07790 histone H2B 30 0.40
At3g49850 MYB -like protein 28 1.5
At1g52620 28 1.5
At5g55920 nucleolar protein-like 27 3.4
At2g43750 cysteine synthase (cpACS1) 27 5.7
At4g26600 unknown protein 26 7.5
At3g59760 cysteine synthase 26 7.5
At5g55280 cell division protein FtsZ chloroplast homolog precurs... 26 9.8
>orf77-01 -chloroplast genome- ribosomal protein S7
Length = 155
Score = 300 bits (768), Expect = 2e-82
Identities = 153/155 (98%), Positives = 155/155 (99%)
Query: 1 MSRRGTAEKKTAKSDPIYRNRLVNMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLS 60
MSRRGTAE+KTAKSDPIYRNRLVNMLVNRILKHGKKSLAYQIIYRA+KKIQQKTETNPLS
Sbjct: 1 MSRRGTAEEKTAKSDPIYRNRLVNMLVNRILKHGKKSLAYQIIYRALKKIQQKTETNPLS 60
Query: 61 VLRQAIRGVTPDIAVKARRVGGSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRNMAFKL 120
VLRQAIRGVTPDIAVKARRVGGSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRNMAFKL
Sbjct: 61 VLRQAIRGVTPDIAVKARRVGGSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRNMAFKL 120
Query: 121 SSELVDAAKGSGDAIRKKEETHRMAEANRAFAHFR 155
SSELVDAAKGSGDAIRKKEETHRMAEANRAFAHFR
Sbjct: 121 SSELVDAAKGSGDAIRKKEETHRMAEANRAFAHFR 155
>rps7 -chloroplast genome- ribosomal protein S7
Length = 155
Score = 300 bits (768), Expect = 2e-82
Identities = 153/155 (98%), Positives = 155/155 (99%)
Query: 1 MSRRGTAEKKTAKSDPIYRNRLVNMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLS 60
MSRRGTAE+KTAKSDPIYRNRLVNMLVNRILKHGKKSLAYQIIYRA+KKIQQKTETNPLS
Sbjct: 1 MSRRGTAEEKTAKSDPIYRNRLVNMLVNRILKHGKKSLAYQIIYRALKKIQQKTETNPLS 60
Query: 61 VLRQAIRGVTPDIAVKARRVGGSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRNMAFKL 120
VLRQAIRGVTPDIAVKARRVGGSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRNMAFKL
Sbjct: 61 VLRQAIRGVTPDIAVKARRVGGSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRNMAFKL 120
Query: 121 SSELVDAAKGSGDAIRKKEETHRMAEANRAFAHFR 155
SSELVDAAKGSGDAIRKKEETHRMAEANRAFAHFR
Sbjct: 121 SSELVDAAKGSGDAIRKKEETHRMAEANRAFAHFR 155
>Atmg01270 -mitochondrial genome- ribosomal protein S7 (RPS7)
Length = 148
Score = 76.3 bits (186), Expect = 6e-15
Identities = 45/140 (32%), Positives = 78/140 (55%), Gaps = 10/140 (7%)
Query: 22 LVNMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLSVLRQAIRGVTPDIAVKARRVG 81
L+ LVN +K GK++ I+Y+ + +TE + + ++ A+ + P V V
Sbjct: 11 LIKKLVNFRMKEGKRTRVRAIVYQTFHR-PARTERDVIKLMVDAVENIKPICEVAKVGVA 69
Query: 82 GSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRNMAFKLS------SELVDAAKGSGDAI 135
G+ + VP + + + LAIRW+L A+ KR +++++S +E++DA + G A
Sbjct: 70 GTIYDVPGIVARDRQQTLAIRWILEAAFKR---RISYRISLEKCSFAEILDAYQKRGSAR 126
Query: 136 RKKEETHRMAEANRAFAHFR 155
RK+E H +A NR+FAHFR
Sbjct: 127 RKRENLHGLASTNRSFAHFR 146
>At2g37270 40S ribosomal protein S5
Length = 207
Score = 60.1 bits (144), Expect = 5e-10
Identities = 48/136 (35%), Positives = 74/136 (54%), Gaps = 9/136 (6%)
Query: 21 RLVNMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLSVLRQAIRGVTPDIAVKARRV 80
RL N L+ +GKK +A +I+ AM+ I ++ NP+ V+ AI P A R+
Sbjct: 74 RLTNSLMMHGRNNGKKLMAVRIVKHAMEIIHLLSDLNPIQVIIDAIVNSGP--REDATRI 131
Query: 81 G--GSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRN---MAFKLSSELVDAAKGSGD-- 133
G G + ++I + AI L +R+ RN +A L+ EL++AAKGS +
Sbjct: 132 GSAGVVRRQAVDISPLRRVNQAIFLLTTGAREAAFRNIKTIAECLADELINAAKGSSNSY 191
Query: 134 AIRKKEETHRMAEANR 149
AI+KK+E R+A+ANR
Sbjct: 192 AIKKKDEIERVAKANR 207
>At3g11940 putative 40S ribosomal protein S5
Length = 207
Score = 59.3 bits (142), Expect = 8e-10
Identities = 47/136 (34%), Positives = 74/136 (53%), Gaps = 9/136 (6%)
Query: 21 RLVNMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLSVLRQAIRGVTPDIAVKARRV 80
RL N L+ +GKK +A +I+ AM+ I ++ NP+ V+ AI P A R+
Sbjct: 74 RLTNSLMMHGRNNGKKLMAVRIVKHAMEIIHLLSDLNPIQVIIDAIVNSGP--REDATRI 131
Query: 81 G--GSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRN---MAFKLSSELVDAAKGSGD-- 133
G G + ++I + AI + +R+ RN +A L+ EL++AAKGS +
Sbjct: 132 GSAGVVRRQAVDISPLRRVNQAIFLITTGAREAAFRNIKTIAECLADELINAAKGSSNSY 191
Query: 134 AIRKKEETHRMAEANR 149
AI+KK+E R+A+ANR
Sbjct: 192 AIKKKDEIERVAKANR 207
>At1g07790 histone H2B
Length = 148
Score = 30.4 bits (67), Expect = 0.40
Identities = 20/76 (26%), Positives = 37/76 (48%), Gaps = 2/76 (2%)
Query: 7 AEKKTAKSDPIYRNRLVNML-VNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLSVLRQA 65
AEKKTA P+ N+ + K GKK + + K+ ++ ET + + +
Sbjct: 10 AEKKTAAERPVEENKAAEKAPAEKKPKAGKKLPPKEAGDKKKKRSKKNVETYKIYIFK-V 68
Query: 66 IRGVTPDIAVKARRVG 81
++ V PDI + ++ +G
Sbjct: 69 LKQVHPDIGISSKAMG 84
>At3g49850 MYB -like protein
Length = 295
Score = 28.5 bits (62), Expect = 1.5
Identities = 17/62 (27%), Positives = 33/62 (52%), Gaps = 1/62 (1%)
Query: 3 RRGTAEKKTAKSDPIYRNRLVNMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLSVL 62
+ GT + +T SDP+Y + ++ N LK ++++ ++ + KK + + PLS
Sbjct: 22 KHGTGKWRTILSDPVY-STILKSRSNVDLKDKWRNISVTALWGSRKKAKLALKRTPLSGS 80
Query: 63 RQ 64
RQ
Sbjct: 81 RQ 82
>At1g52620
Length = 819
Score = 28.5 bits (62), Expect = 1.5
Identities = 22/67 (32%), Positives = 29/67 (42%), Gaps = 17/67 (25%)
Query: 24 NMLVNRILKHGKKSLAYQIIYRAMKK--IQQKTETNPL---------------SVLRQAI 66
N+L+NR+ K GKK +A + A KK I PL +L+ A
Sbjct: 349 NILINRLCKEGKKEVAVGFLDEASKKGLIPNNLSYAPLIQAYCKSKEYDIASKLLLQMAE 408
Query: 67 RGVTPDI 73
RG PDI
Sbjct: 409 RGCKPDI 415
>At5g55920 nucleolar protein-like
Length = 682
Score = 27.3 bits (59), Expect = 3.4
Identities = 26/98 (26%), Positives = 39/98 (39%), Gaps = 6/98 (6%)
Query: 41 QIIYRAMKKIQQKTETNPLSVLRQAI------RGVTPDIAVKARRVGGSTHQVPIEIGST 94
++I K+ TN L R+ + RGV D K +VG + + IG+T
Sbjct: 274 ELIEAFEKQRPTSIRTNTLKTRRRDLADVLLNRGVNLDPLSKWSKVGLVIYDSQVPIGAT 333
Query: 95 QGKALAIRWLLGASRKRPGRNMAFKLSSELVDAAKGSG 132
L GAS P +A + + +VD A G
Sbjct: 334 PEYLAGYYMLQGASSFLPVMALAPRENERIVDVAAAPG 371
>At2g43750 cysteine synthase (cpACS1)
Length = 392
Score = 26.6 bits (57), Expect = 5.7
Identities = 21/72 (29%), Positives = 31/72 (42%)
Query: 69 VTPDIAVKARRVGGSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRNMAFKLSSELVDAA 128
+TP +V G+T I +++G L + S +R AF L + A
Sbjct: 133 ITPGKSVLVESTSGNTGIGLAFIAASKGYKLILTMPASMSLERRVLLRAFGAELVLTEPA 192
Query: 129 KGSGDAIRKKEE 140
KG AI+K EE
Sbjct: 193 KGMTGAIQKAEE 204
>At4g26600 unknown protein
Length = 671
Score = 26.2 bits (56), Expect = 7.5
Identities = 26/98 (26%), Positives = 37/98 (37%), Gaps = 6/98 (6%)
Query: 41 QIIYRAMKKIQQKTETNPLSVLRQAI------RGVTPDIAVKARRVGGSTHQVPIEIGST 94
++I KK TN L R+ + RGV D K +VG + + IG+T
Sbjct: 257 ELIEAFEKKRPTSIRTNTLKTRRRDLADILLNRGVNLDPLSKWSKVGLIVYDSQVPIGAT 316
Query: 95 QGKALAIRWLLGASRKRPGRNMAFKLSSELVDAAKGSG 132
L AS P +A + +VD A G
Sbjct: 317 PEYLAGFYMLQSASSFLPVMALAPREKERVVDMAAAPG 354
>At3g59760 cysteine synthase
Length = 430
Score = 26.2 bits (56), Expect = 7.5
Identities = 20/72 (27%), Positives = 31/72 (42%)
Query: 69 VTPDIAVKARRVGGSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRNMAFKLSSELVDAA 128
++P +V G+T I +++G L + S +R AF L D A
Sbjct: 171 ISPGKSVLVEPTSGNTGIGLAFIAASRGYRLILTMPASMSMERRVLLKAFGAELVLTDPA 230
Query: 129 KGSGDAIRKKEE 140
KG A++K EE
Sbjct: 231 KGMTGAVQKAEE 242
>At5g55280 cell division protein FtsZ chloroplast homolog precursor
(sp|Q42545)
Length = 433
Score = 25.8 bits (55), Expect = 9.8
Identities = 13/46 (28%), Positives = 23/46 (49%)
Query: 24 NMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLSVLRQAIRGV 69
N VNR++ G +S+ + I + + Q + NPL + RG+
Sbjct: 86 NNAVNRMISSGLQSVDFYAINTDSQALLQSSAENPLQIGELLTRGL 131
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.131 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,943,222
Number of Sequences: 26719
Number of extensions: 102421
Number of successful extensions: 309
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 293
Number of HSP's gapped (non-prelim): 13
length of query: 155
length of database: 11,318,596
effective HSP length: 91
effective length of query: 64
effective length of database: 8,887,167
effective search space: 568778688
effective search space used: 568778688
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Description of BAB33240.1

