
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
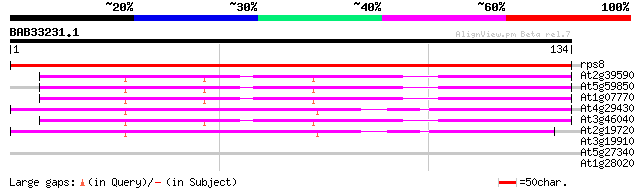
Query= BAB33231.1 134 aa
(134 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
rps8 -chloroplast genome- ribosomal protein S8 219 4e-58
At2g39590 40S ribosomal protein S15A 44 2e-05
At5g59850 cytoplasmic ribosomal protein S15a - like 43 6e-05
At1g07770 Putative 40S ribosomal protein S15A 43 6e-05
At4g29430 ribosomal protein S15a homolog 42 1e-04
At3g46040 cytoplasmic ribosomal protein S15a -like 42 1e-04
At2g19720 40S ribosomal protein S15A 42 1e-04
At3g19910 unknown protein 28 1.9
At5g27340 hypothetical protein 26 5.4
At1g28020 hypothetical protein 25 9.3
>rps8 -chloroplast genome- ribosomal protein S8
Length = 134
Score = 219 bits (557), Expect = 4e-58
Identities = 110/134 (82%), Positives = 123/134 (91%)
Query: 1 MGKDTIADIITSIRNADMNRKRTVQIPFTNITENTVKILLREGFIENVRKHRESDKSFLV 60
MGKDTIADIITSIRNADMNRK TV+I TNITE+ VKILLREGFIENVRKHRE+++ FL+
Sbjct: 1 MGKDTIADIITSIRNADMNRKGTVRIGSTNITESIVKILLREGFIENVRKHRENNQYFLI 60
Query: 61 LTLRYRRNTKGSYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREA 120
LTLR+RRN K SYKT LNLKRIS PGLRIY N Q+IPRILGG+GIVILSTS+GIMTDREA
Sbjct: 61 LTLRHRRNKKESYKTILNLKRISRPGLRIYSNSQRIPRILGGIGIVILSTSQGIMTDREA 120
Query: 121 RLEKIGGEVLCYIW 134
RL++IGGE+LCYIW
Sbjct: 121 RLKRIGGEILCYIW 134
>At2g39590 40S ribosomal protein S15A
Length = 136
Score = 44.3 bits (103), Expect = 2e-05
Identities = 39/133 (29%), Positives = 64/133 (47%), Gaps = 17/133 (12%)
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + S+ NA+ KR V I P + + + ++ + G+I E V HR +V+ L
Sbjct: 15 DCLKSMSNAEKQGKRQVMIRPSSKVIIKFLTVMQKHGYIAEFEYVDDHRSGK---IVVEL 71
Query: 64 RYRRNTKG--SYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREAR 121
R N G S + L +K I ++ + Q G ++L+TS GIM EAR
Sbjct: 72 NGRLNNCGVISPRYDLGVKEIEGWTAKLLPSRQ--------FGYIVLTTSAGIMDHEEAR 123
Query: 122 LEKIGGEVLCYIW 134
+ +GG+VL + +
Sbjct: 124 KKNVGGKVLGFFY 136
>At5g59850 cytoplasmic ribosomal protein S15a - like
Length = 130
Score = 42.7 bits (99), Expect = 6e-05
Identities = 39/133 (29%), Positives = 64/133 (47%), Gaps = 17/133 (12%)
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + S+ NA+ KR V I P + + + ++ + G+I E V HR +V+ L
Sbjct: 9 DALKSMYNAEKRGKRQVMIRPSSKVIIKFLIVMQKHGYIGEFEYVDDHRSGK---IVVEL 65
Query: 64 RYRRNTKG--SYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREAR 121
R N G S + + +K I R+ + Q G ++L+TS GIM EAR
Sbjct: 66 NGRLNKCGVISPRFDVGVKEIEGWTARLLPSRQ--------FGYIVLTTSAGIMDHEEAR 117
Query: 122 LEKIGGEVLCYIW 134
+ +GG+VL + +
Sbjct: 118 RKNVGGKVLGFFY 130
>At1g07770 Putative 40S ribosomal protein S15A
Length = 130
Score = 42.7 bits (99), Expect = 6e-05
Identities = 39/133 (29%), Positives = 64/133 (47%), Gaps = 17/133 (12%)
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + S+ NA+ KR V I P + + + ++ + G+I E V HR +V+ L
Sbjct: 9 DALKSMYNAEKRGKRQVMIRPSSKVIIKFLIVMQKHGYIGEFEYVDDHRSGK---IVVEL 65
Query: 64 RYRRNTKG--SYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREAR 121
R N G S + + +K I R+ + Q G ++L+TS GIM EAR
Sbjct: 66 NGRLNKCGVISPRFDVGVKEIEGWTARLLPSRQ--------FGYIVLTTSAGIMDHEEAR 117
Query: 122 LEKIGGEVLCYIW 134
+ +GG+VL + +
Sbjct: 118 RKNVGGKVLGFFY 130
>At4g29430 ribosomal protein S15a homolog
Length = 129
Score = 42.0 bits (97), Expect = 1e-04
Identities = 32/137 (23%), Positives = 67/137 (48%), Gaps = 11/137 (8%)
Query: 1 MGKDTIADIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFIENVRKHRESDKSFL 59
MG+ + D + +I NA+ K +V++ P + + + ++I+ +G+I+N + + +
Sbjct: 1 MGRRILNDALRTIVNAERRGKASVELKPISTVMSSFLRIMKEKGYIKNFQVYDPHRVGRI 60
Query: 60 VLTLRYRRNTKGS--YKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTD 117
+ L+ R N + Y+ + K I Y + +P G V+++T GI+
Sbjct: 61 TVDLQGRVNDCKALTYRQDVRAKEIEK------YTERTLPT--RQWGYVVITTPDGILDH 112
Query: 118 REARLEKIGGEVLCYIW 134
EA +GG+VL + +
Sbjct: 113 EEAIKRNVGGQVLGFFY 129
>At3g46040 cytoplasmic ribosomal protein S15a -like
Length = 130
Score = 42.0 bits (97), Expect = 1e-04
Identities = 39/133 (29%), Positives = 64/133 (47%), Gaps = 17/133 (12%)
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + S+ NA+ KR V I P + + + ++ + G+I E V HR +V+ L
Sbjct: 9 DGLKSMYNAEKRGKRQVMIRPSSKVIIKFLIVMQKHGYIGEFEYVDDHRSGK---IVVEL 65
Query: 64 RYRRNTKG--SYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREAR 121
R N G S + + +K I R+ + Q G ++L+TS GIM EAR
Sbjct: 66 NGRLNKCGVISPRFDVGVKEIEGWTARLLPSRQ--------FGFIVLTTSAGIMDHEEAR 117
Query: 122 LEKIGGEVLCYIW 134
+ +GG+VL + +
Sbjct: 118 RKNVGGKVLGFFY 130
>At2g19720 40S ribosomal protein S15A
Length = 129
Score = 42.0 bits (97), Expect = 1e-04
Identities = 32/133 (24%), Positives = 64/133 (48%), Gaps = 11/133 (8%)
Query: 1 MGKDTIADIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFIENVRKHRESDKSFL 59
MG+ + D + +I NA+ K +V++ P + + + +KI+ +G+I+N + H +
Sbjct: 1 MGRRILNDALRTIVNAEKRGKASVELKPVSTVMSSFLKIMKEKGYIKNFQVHDPHRVGRI 60
Query: 60 VLTLRYRRNTKGS--YKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTD 117
+ L+ R N + Y+ + I Y + +P G ++++T GI+
Sbjct: 61 TVDLQGRVNDCKALTYRQDVKANEIGQ------YTERTLPT--RQWGYIVITTPDGILDH 112
Query: 118 REARLEKIGGEVL 130
EA +GG+VL
Sbjct: 113 EEAIKRNVGGQVL 125
>At3g19910 unknown protein
Length = 340
Score = 27.7 bits (60), Expect = 1.9
Identities = 13/56 (23%), Positives = 31/56 (55%), Gaps = 7/56 (12%)
Query: 20 RKRTVQIPFTNITENTVKILLREGFIENVRKHRESDKSFLVLTLRYRRNTKGSYKT 75
R + ++PFTN+++ + L R +E ++++++LT+ + GS++T
Sbjct: 80 RSSSARVPFTNLSQIDADLAL-------ARTLQEQERAYMMLTMNSEISDYGSWET 128
>At5g27340 hypothetical protein
Length = 238
Score = 26.2 bits (56), Expect = 5.4
Identities = 15/55 (27%), Positives = 27/55 (48%)
Query: 46 ENVRKHRESDKSFLVLTLRYRRNTKGSYKTFLNLKRISTPGLRIYYNYQKIPRIL 100
E+VR+ E +SF ++ K Y+ L+ K I + L + Y Y++ +L
Sbjct: 62 EHVRQVWEIGRSFRERSMELLLGIKQPYQRQLSTKEIISSSLELRYKYEEFDDLL 116
>At1g28020 hypothetical protein
Length = 612
Score = 25.4 bits (54), Expect = 9.3
Identities = 15/58 (25%), Positives = 26/58 (43%)
Query: 30 NITENTVKILLREGFIENVRKHRESDKSFLVLTLRYRRNTKGSYKTFLNLKRISTPGL 87
++ EN V + E F E++ K+ D + L Y R+ K K +++ GL
Sbjct: 112 HLIENVVGLEEAEKFFESIPKNARGDSVYTSLLNSYARSDKTLCKAEATFQKMRDLGL 169
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.141 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,738,699
Number of Sequences: 26719
Number of extensions: 103787
Number of successful extensions: 197
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 186
Number of HSP's gapped (non-prelim): 11
length of query: 134
length of database: 11,318,596
effective HSP length: 88
effective length of query: 46
effective length of database: 8,967,324
effective search space: 412496904
effective search space used: 412496904
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 54 (25.4 bits)
Description of BAB33231.1

