
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
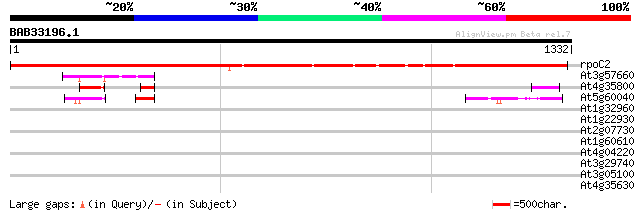
Query= BAB33196.1 1332 aa
(1332 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
rpoC2 -chloroplast genome- RNA polymerase beta' subunit-2 1921 0.0
At3g57660 DNA-directed RNA polymerase I 190K chain - like protein 77 5e-14
At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain 51 4e-06
At5g60040 DNA-directed RNA polymerase - like protein 50 9e-06
At1g32960 unknown protein 32 1.9
At1g22930 unknown protein 32 2.4
At2g07730 putative non-LTR retroelement reverse transcriptase 32 3.2
At1g60610 unknown protein 32 3.2
At4g04220 putative disease resistance protein, predicted GPI-anc... 31 4.2
At3g29740 hypothetical protein 31 4.2
At3g05100 unknown protein 31 4.2
At4g35630 phosphoserine aminotransferase 30 7.1
>rpoC2 -chloroplast genome- RNA polymerase beta' subunit-2
Length = 1376
Score = 1921 bits (4976), Expect = 0.0
Identities = 1001/1358 (73%), Positives = 1125/1358 (82%), Gaps = 53/1358 (3%)
Query: 1 MAERANLVFHNKVIGGTAIKRIISRLIDHFGMAYTSHILDQVKTLGFRQATTTSISLGID 60
MAERANLVFHNKVI GTAIKR+ISRLIDHFGMAYTSHILDQVKTLGF+QAT TSISLGID
Sbjct: 1 MAERANLVFHNKVIDGTAIKRLISRLIDHFGMAYTSHILDQVKTLGFQQATATSISLGID 60
Query: 61 DLLTIPSKGWLVQDAEQQSLILEKHHHYGNVHAVEKLRQSIEIWYATSEYLRQEMNPNFS 120
DLLTIPSKGWLVQDAEQQS ILEKHHHYGNVHAVEKLRQSIEIWYATSEYLRQEMNPNF
Sbjct: 61 DLLTIPSKGWLVQDAEQQSWILEKHHHYGNVHAVEKLRQSIEIWYATSEYLRQEMNPNFR 120
Query: 121 MTDPFNPVHIMSFSGARGNASQVHQLVGMRGLMSDPQGQMIDLPIQSNLREGLSLTEYII 180
MTDPFNPVH+MSFSGARGNASQVHQLVGMRGLMSDPQGQMIDLPIQSNLREGLSLTEYII
Sbjct: 121 MTDPFNPVHMMSFSGARGNASQVHQLVGMRGLMSDPQGQMIDLPIQSNLREGLSLTEYII 180
Query: 181 SCYGARKGVVDTAVRTSDAGYLTRRLVEVVQHIVVRRTDCGTVRGISVNTRN--RMMSER 238
SCYGARKGVVDTAVRTSDAGYLTRRLVEVVQHIVVRRTDCGT+RGISV+ RN RMMSER
Sbjct: 181 SCYGARKGVVDTAVRTSDAGYLTRRLVEVVQHIVVRRTDCGTIRGISVSPRNKNRMMSER 240
Query: 239 ILIQTLIGRVLADDIYIGSRCIVVRNQDIGIGLINRFINFQTQPIFIRTPFTCRNTSWIC 298
I IQTLIGRVLADDIYIGSRC+ RNQD+GIGL+NR I F TQ I IRTPFTCR+TSWIC
Sbjct: 241 IFIQTLIGRVLADDIYIGSRCVAFRNQDLGIGLVNRLITFGTQSISIRTPFTCRSTSWIC 300
Query: 299 RLCYGRSPIHGNLVELGEAVGIIAGQSIGEPGTQLTLRTFHTGGVFTGGTAEYVRSPSNG 358
RLCYGRSP HG+LVELGEAVGIIAGQSIGEPGTQLTLRTFHTGGVFTGGTAE+VR+P NG
Sbjct: 301 RLCYGRSPTHGDLVELGEAVGIIAGQSIGEPGTQLTLRTFHTGGVFTGGTAEHVRAPYNG 360
Query: 359 KIKFNENSAYPTRTRHGHPAFLCYIDLYVTIESNDIMHNVIIPPKSFLLVQNDQYVKSEQ 418
KIKFNE+ +PTRTRHGHPAFLCYIDL V IES DI+H+V IPPKSFLLVQNDQYV+SEQ
Sbjct: 361 KIKFNEDLVHPTRTRHGHPAFLCYIDLSVIIESEDIIHSVTIPPKSFLLVQNDQYVESEQ 420
Query: 419 IIAEIRAGTYTLNLKEKVRKHIFSDSEGEMHWSTNIYHVSEFAYSNVHILPKTSHLWILS 478
+IAEIR GTYT + KE+VRK+I+SDSEGEMHWST++ H EF YSNVH+LPKTSHLWILS
Sbjct: 421 VIAEIREGTYTFHFKERVRKYIYSDSEGEMHWSTDVSHAPEFTYSNVHLLPKTSHLWILS 480
Query: 479 GNSHKSDTVSLSLLKDQDQMSTHSLPTAKRNTSNFLVSNNQV------------------ 520
G S S + S+ KDQDQM+ L +++ S+ V+N+QV
Sbjct: 481 GGSCGSSLIRFSIHKDQDQMNIPFLSAERKSISSLSVNNDQVSQKFFSSDFADPKKLGIY 540
Query: 521 -------RLCPDHCHFMHPTISPDTSNLLAKKRRNRFIIPFLFRSIRERNNELMPD--IS 571
L H + ++ I + S+LLAK+RRNRF+IP F+SI+E+ E +P IS
Sbjct: 541 DYSELNGNLGTSHYNLIYSAIFHENSDLLAKRRRNRFLIP--FQSIQEQEKEFIPQSGIS 598
Query: 572 VEIPIDGIIHKNSILAYFDDPQYRTQSSGIAKYKTIGIHSIFQKEDLIEYRGIREFKPKY 631
VEIPI+GI +NSI A+FDDP+YR +SSGI KY T+ SI QKED+IEYRG+++ K KY
Sbjct: 599 VEIPINGIFRRNSIFAFFDDPRYRRKSSGILKYGTLKADSIIQKEDMIEYRGVQKIKTKY 658
Query: 632 QIKVDRFFFIPQEVHILSESSSIMVRNNSIIGVNTPITLNKKSRVGGLVRVEKNKKKIEL 691
++KVDRFFFIP+EVHIL ESS+IMV+N SIIGV+T +TLN +S+VGGL+RVEK KK+IEL
Sbjct: 659 EMKVDRFFFIPEEVHILPESSAIMVQNYSIIGVDTRLTLNIRSQVGGLIRVEKKKKRIEL 718
Query: 692 KIFSGDIHFPGEIDKISQHSAILIPPEMVKKKNSKESKKKTNWRYIQWITTTKKKYFVLV 751
KIFSGDIHFP + DKIS+HS ILIPP KKNSKESKK NW Y+Q IT TKKK+FVLV
Sbjct: 719 KIFSGDIHFPDKTDKISRHSGILIPPGR-GKKNSKESKKFKNWIYVQRITPTKKKFFVLV 777
Query: 752 RPVILYDIADSINLVKLFPQDLFKEWDNLELKVLNFILYGNGKSIRGILDTSIQLVRTCL 811
RPV Y+IADSINL LFPQDLF+E DN++L+V N+ILYGNGK RGI DTSIQLVRTCL
Sbjct: 778 RPVATYEIADSINLATLFPQDLFREKDNIQLRVFNYILYGNGKPTRGISDTSIQLVRTCL 837
Query: 812 VLNWNEDEKSSSIEEALASFVEVSTNGLIRYFLRIDLVKSHISYIRKRNDPSSSGLISYN 871
VLNW +K+SS+EE A FVEVST GLI+ F+RI LVKSHISYIRKRN+ SGLIS +
Sbjct: 838 VLNW---DKNSSLEEVRAFFVEVSTKGLIQDFIRIGLVKSHISYIRKRNNSPDSGLISAD 894
Query: 872 ESDRININPFFSIYKEN--IQQSLSQKHGTIRMLLNRNKENRSFIILSSSNCFQMGPFNN 929
++NPF+SI ++ +QQSL Q HGTIRM LNRNKE++S +ILSSSNCF+MGPFN+
Sbjct: 895 -----HMNPFYSISPKSGILQQSLRQNHGTIRMFLNRNKESQSLLILSSSNCFRMGPFNH 949
Query: 930 VKYHNGIKEEINQFKRNHKIPIKISLGPLGVAPQIANFFSFYHLITHNKISSIKKNLQLN 989
VK+HN I + I K+N I IK S GPLG A I+NF+SF L+T+N+IS I K QL+
Sbjct: 950 VKHHNVINQSI---KKNTLITIKNSSGPLGTATPISNFYSFLPLLTYNQISLI-KYFQLD 1005
Query: 990 KFKETFQVIKYYLMDENERIYKPDLYNNIILNPFHLNWDFIHP----NYCEKTFPIISLG 1045
K FQ I YL+DEN I D Y+N++LNPF LNW F+H NYCE+T IISLG
Sbjct: 1006 NLKYIFQKINSYLIDENGIILNLDPYSNVVLNPFKLNWYFLHQNYHHNYCEETSTIISLG 1065
Query: 1046 QFICENVCIVQTKNGPNLKSGQVITVQMDFVGIRLANPYLATPGATIHGHYGEMLYEGDI 1105
QF CENVCI K P+LKSGQV+ VQ D IR A PYLATPGA +HGHY E+LYEGD
Sbjct: 1066 QFFCENVCI--AKKEPHLKSGQVLIVQRDSAVIRSAKPYLATPGAKVHGHYSEILYEGDT 1123
Query: 1106 LVTFIYEKSRSGDITQGLPKVEQVLEVRSIDSISMNLEKRIDAWNERITGILGIPWRFLI 1165
LVTFIYEKSRSGDITQGLPKVEQVLEVRSIDSIS+NLEKRI WN+ IT ILGIPW FLI
Sbjct: 1124 LVTFIYEKSRSGDITQGLPKVEQVLEVRSIDSISLNLEKRIKGWNKCITRILGIPWGFLI 1183
Query: 1166 GAELTIAQSRISLVNKIQRVYRSQGVHIHNRHIEIIVRQITSKVLVSEDGMSNVFSPGEL 1225
GAELTI QSRISLVNKIQ+VYRSQGV IHNRHIEIIVRQITSKVLVSE+GMSNVF PGEL
Sbjct: 1184 GAELTIVQSRISLVNKIQKVYRSQGVQIHNRHIEIIVRQITSKVLVSEEGMSNVFLPGEL 1243
Query: 1226 IGLLRAQRTGRALEESICYRTLLLGITKTSLNTQSFISEASFQETARVLAKAALRGRIDW 1285
IGLLRA+RTGRALEE+ICYR +LLGIT+ SLNTQSFISEASFQETARVLAKAALRGRIDW
Sbjct: 1244 IGLLRAERTGRALEEAICYRAVLLGITRASLNTQSFISEASFQETARVLAKAALRGRIDW 1303
Query: 1286 LKGLKENLVLGGIIPVGTGFKKIGDRSSARQDTKITLE 1323
LKGLKEN+VLGG+IP GTGF K G +RQ T I LE
Sbjct: 1304 LKGLKENVVLGGVIPAGTGFNK-GLVHCSRQHTNIILE 1340
>At3g57660 DNA-directed RNA polymerase I 190K chain - like protein
Length = 1670
Score = 77.4 bits (189), Expect = 5e-14
Identities = 72/254 (28%), Positives = 114/254 (44%), Gaps = 44/254 (17%)
Query: 126 NPVHIMSFSGARG---NASQVHQLVGMRGLMSDPQGQMID---LP--------------I 165
N + +M+ SGA+G N Q+ +G + L +M+ LP I
Sbjct: 936 NCISLMTISGAKGSKVNFQQISSHLGQQDLEGKRVPRMVSGKTLPCFHPWDWSPRAGGFI 995
Query: 166 QSNLREGLSLTEYIISCYGARKGVVDTAVRTSDAGYLTRRLVEVVQHIVVRRTDCGTVR- 224
GL EY C R+G+VDTAV+TS +GYL R L++ ++ + V DC TVR
Sbjct: 996 SDRFLSGLRPQEYYFHCMAGREGLVDTAVKTSRSGYLQRCLMKNLESLKVNY-DC-TVRD 1053
Query: 225 -------------GISVNTRN--RMMSERILIQTLIGRVLADDIYIGSRCIVVRNQDIGI 269
G+ V+ + E + Q ++ + ++D+ G+ + D+ I
Sbjct: 1054 ADGSIIQFQYGEDGVDVHRSSFIEKFKELTINQDMVLQKCSEDMLSGASSYI---SDLPI 1110
Query: 270 GLINRFINF-QTQPIFIRTPFTCRNTSWICRLCYGRSPIHGNLVELGEAVGIIAGQSIGE 328
L F + P+ R + +L +S +L + GE VG++A QS+GE
Sbjct: 1111 SLKKGAEKFVEAMPMNERIASKFVRQEELLKLV--KSKFFASLAQPGEPVGVLAAQSVGE 1168
Query: 329 PGTQLTLRTFHTGG 342
P TQ+TL TFH G
Sbjct: 1169 PSTQMTLNTFHLAG 1182
>At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain
Length = 1840
Score = 51.2 bits (121), Expect = 4e-06
Identities = 24/60 (40%), Positives = 41/60 (68%), Gaps = 2/60 (3%)
Query: 165 IQSNLREGLSLTEYIISCYGARKGVVDTAVRTSDAGYLTRRLVEVVQHIVVRRTDCGTVR 224
++++ GL+ E+ G R+G++DTAV+TS+ GY+ RRLV+ ++ I+V+ GTVR
Sbjct: 820 VENSYLRGLTPQEFFFHAMGGREGLIDTAVKTSETGYIQRRLVKAMEDIMVKYD--GTVR 877
Score = 48.1 bits (113), Expect = 3e-05
Identities = 24/67 (35%), Positives = 39/67 (57%), Gaps = 2/67 (2%)
Query: 1240 ESICYRTLLLGITKTSLNTQSF--ISEASFQETARVLAKAALRGRIDWLKGLKENLVLGG 1297
+++ YR L+ IT+ +N + SF+ET +L AA D L+G+ EN++LG
Sbjct: 1395 DTMTYRGHLMAITRHGINRNDTGPLMRCSFEETVDILLDAAAYAETDCLRGVTENIMLGQ 1454
Query: 1298 IIPVGTG 1304
+ P+GTG
Sbjct: 1455 LAPIGTG 1461
Score = 46.6 bits (109), Expect = 1e-04
Identities = 21/34 (61%), Positives = 25/34 (72%)
Query: 310 NLVELGEAVGIIAGQSIGEPGTQLTLRTFHTGGV 343
+LV GE +G +A QSIGEP TQ+TL TFH GV
Sbjct: 1077 SLVAPGEMIGCVAAQSIGEPATQMTLNTFHYAGV 1110
>At5g60040 DNA-directed RNA polymerase - like protein
Length = 1328
Score = 50.1 bits (118), Expect = 9e-06
Identities = 23/45 (51%), Positives = 30/45 (66%)
Query: 299 RLCYGRSPIHGNLVELGEAVGIIAGQSIGEPGTQLTLRTFHTGGV 343
++ Y S + +E G A+G I QSIGEPGTQ+TL+TFH GV
Sbjct: 965 QVLYKASGVTDKQLEAGTAIGTIGAQSIGEPGTQMTLKTFHFAGV 1009
Score = 49.7 bits (117), Expect = 1e-05
Identities = 37/119 (31%), Positives = 61/119 (51%), Gaps = 24/119 (20%)
Query: 130 IMSFSGARGNASQVHQLVGMRGLMS-----DPQGQMID--LP--------------IQSN 168
IMS G++G+ + Q+V G + P G ID LP + ++
Sbjct: 756 IMSQCGSKGSPINISQMVACVGQQTVNGHRAPDG-FIDRSLPHFPRMSKSPAAKGFVANS 814
Query: 169 LREGLSLTEYIISCYGARKGVVDTAVRTSDAGYLTRRLVEVVQHIVVRRTDCGTVRGIS 227
GL+ TE+ G R+G+VDTAV+T+ GY++RRL++ ++ ++V + TVR S
Sbjct: 815 FYSGLTATEFFFHTMGGREGLVDTAVKTASTGYMSRRLMKALEDLLVHYDN--TVRNAS 871
Score = 47.0 bits (110), Expect = 7e-05
Identities = 59/253 (23%), Positives = 111/253 (43%), Gaps = 63/253 (24%)
Query: 1082 NPYLATPGATIHGHYGEMLYEGDILVTFIYEKSRSG----DITQGLPKVEQVLEVRSIDS 1137
N L TP ++ + +L G + +T + +KSR+ ++ G+ VE+V+ +D
Sbjct: 1102 NSILKTPRIKLNDNDIRVLDTG-LDITPVVDKSRAHFNLHNLKNGIKTVERVVVAEDMDK 1160
Query: 1138 ISMNLEKRIDA---WNERITG-----ILGIPWR----------FLIGAELTIAQSRISLV 1179
K+ID W + G ++G P + L I +R +++
Sbjct: 1161 -----SKQIDGKTKWKLFVEGTNLLAVMGTPGINGRTTTSNNVVEVSKTLGIEAARTTII 1215
Query: 1180 NKIQRVYRSQGVHIHNRHIEIIVRQITSKVLVSEDGMSNVFSPGELIGLLRAQRTGRALE 1239
++I V + G+ I RH+ ++ +T + GE++G+ QRTG
Sbjct: 1216 DEIGTVMGNHGMSIDIRHMMLLADVMTYR--------------GEVLGI---QRTG---- 1254
Query: 1240 ESICYRTLLLGITKTSLNTQSFISEASFQETARVLAKAALRGRIDWLKGLKENLVLGGII 1299
I K +S + +ASF+ T L AA G++D ++G+ E +++G +
Sbjct: 1255 -----------IQKMD---KSVLMQASFERTGDHLFSAAASGKVDNIEGVTECVIMGIPM 1300
Query: 1300 PVGTGFKKIGDRS 1312
+GTG K+ R+
Sbjct: 1301 KLGTGILKVLQRT 1313
>At1g32960 unknown protein
Length = 777
Score = 32.3 bits (72), Expect = 1.9
Identities = 26/110 (23%), Positives = 53/110 (47%), Gaps = 9/110 (8%)
Query: 129 HIMSFSGARGNASQVHQLVGMRGLMSDPQGQMIDLPIQS----NLREGLSLTEYIISCYG 184
+I+ A N S + QLVG + S+P+ ++D+ + S NL++ ++LT + +
Sbjct: 637 YILYLCSAGYNDSSISQLVGQITVCSNPKPSVLDVNLPSITIPNLKDEVTLTRTVTNV-- 694
Query: 185 ARKGVVDTAVRTSDAGYLTRRLVEVVQHIVVRRTDCGTVRGISVNTRNRM 234
G+VD+ + S L R+V + +V + V+T +++
Sbjct: 695 ---GLVDSVYKVSVEPPLGVRVVVTPETLVFNSKTISVSFTVRVSTTHKI 741
>At1g22930 unknown protein
Length = 1131
Score = 32.0 bits (71), Expect = 2.4
Identities = 23/91 (25%), Positives = 40/91 (43%), Gaps = 7/91 (7%)
Query: 58 GIDDLLTIPSKGWLVQDAEQQSLILEKHHHYGNVHAVEKLRQSIEIWYATSEYLRQEMNP 117
G+D +LT K +Q + Q L+ EK H V VE++ ++ +++ +P
Sbjct: 526 GVDTMLTHDKKAIQMQVTQDQELLTEKVRHLSGVAGVERMESALLETRTKYFQAKEDGSP 585
Query: 118 N-------FSMTDPFNPVHIMSFSGARGNAS 141
FS + +PV +S S +R S
Sbjct: 586 MANQLAHFFSPSPASSPVQSVSSSSSRSKDS 616
>At2g07730 putative non-LTR retroelement reverse transcriptase
Length = 970
Score = 31.6 bits (70), Expect = 3.2
Identities = 26/107 (24%), Positives = 46/107 (42%), Gaps = 12/107 (11%)
Query: 393 DIMHNVIIPPKSFLLVQNDQYVKSEQIIAEIRAGTYTLNLKEKVRKHIFSDSEGEMHWST 452
D+MH+ LL Q + +K ++ E + +EK R I S +G+ W T
Sbjct: 72 DVMHS------DELLAQEEILLKDLDLVLEQEETLWFQKSREKKRNRIESPKDGDNRWMT 125
Query: 453 NIYHVSEFA---YSNVHILPKTSHLW---ILSGNSHKSDTVSLSLLK 493
+ + A Y+ ++ L S +W + G + +D +LLK
Sbjct: 126 DKVELETMALNYYTRLYSLEDVSAVWNMLPIGGFATLTDGEKAALLK 172
>At1g60610 unknown protein
Length = 340
Score = 31.6 bits (70), Expect = 3.2
Identities = 24/107 (22%), Positives = 55/107 (50%), Gaps = 7/107 (6%)
Query: 806 LVRTCLVLNWNEDEKSSSIEEALASFVEVSTNGLIRYFLRIDLVKSH---ISYIRKRNDP 862
LV T L L++++DE++SS+ A S + + +R+DL + + +I+ R D
Sbjct: 104 LVSTGLRLSYDDDERNSSVTSANGS-ITTPVYQSLGDNIRLDLNRQNDELDQFIKFRADQ 162
Query: 863 SSSGLISYNESDRININPFFSIYKENIQQSLSQKHGTIRMLLNRNKE 909
+ G+ + + ++ F + ++++ + L +K I + +N+E
Sbjct: 163 MAKGV---RDIKQRHVTSFVTALEKDVSKKLQEKDHEIESMNKKNRE 206
>At4g04220 putative disease resistance protein, predicted
GPI-anchored protein
Length = 766
Score = 31.2 bits (69), Expect = 4.2
Identities = 35/145 (24%), Positives = 66/145 (45%), Gaps = 17/145 (11%)
Query: 758 DIADSINLVKLFPQDLFK---EWDNLELKVL--NFILYG----NGKSIRGILDTSIQLVR 808
DI + L+++ +D+F W N + + NF LY + + G + TS+ ++
Sbjct: 562 DIPNIERLIEIESEDIFSLVVNWKNSKQVLFDRNFYLYTLLDLSKNKLHGEIPTSLGNLK 621
Query: 809 TCLVLNWNEDEKSSSIEEALASFVEVSTNGLIRYFLRIDLVK-----SHISYIRKRNDPS 863
+ VLN + +E S I ++ +V + L L ++ K S ++ + RN+
Sbjct: 622 SLKVLNLSNNEFSGLIPQSFGDLEKVESLDLSHNNLTGEIPKTLSKLSELNTLDLRNNKL 681
Query: 864 SSGLISYNESDRININPFFSIYKEN 888
+ + DR+N NP +IY N
Sbjct: 682 KGRIPESPQLDRLN-NP--NIYANN 703
>At3g29740 hypothetical protein
Length = 87
Score = 31.2 bits (69), Expect = 4.2
Identities = 25/83 (30%), Positives = 43/83 (51%), Gaps = 7/83 (8%)
Query: 671 NKKSRVGGLVRVEKNKKKIELKIFSGDIHFPGEIDKISQHSAILIPPEMVKKK--NSKES 728
NK+ VGGL R+ K ++++++++ +GD G IS H +L V K ++E
Sbjct: 6 NKELFVGGLARILKEQRQVDVRLKAGDSDQKGV--SISAHKLVLSARSEVFKMILETEEI 63
Query: 729 KKKTNWRYIQWITTTKKKYFVLV 751
K T + IT ++ K+ LV
Sbjct: 64 KATTT---LDTITLSELKHTELV 83
>At3g05100 unknown protein
Length = 336
Score = 31.2 bits (69), Expect = 4.2
Identities = 17/65 (26%), Positives = 31/65 (47%), Gaps = 1/65 (1%)
Query: 549 RFIIPFLFRSIRERNNELMPDISVEIPIDGIIHKNSILAYFDDPQYRTQSSGIAKYKTIG 608
R++ P F + + LM + E+P G++HK ++ DD ++ S A Y I
Sbjct: 194 RYLSPQHFHCLEKDELSLMAALRYELPSQGLLHKRPLIVRGDDMEFSKFGSDTA-YDLIY 252
Query: 609 IHSIF 613
++F
Sbjct: 253 ASAVF 257
>At4g35630 phosphoserine aminotransferase
Length = 430
Score = 30.4 bits (67), Expect = 7.1
Identities = 21/80 (26%), Positives = 36/80 (44%), Gaps = 9/80 (11%)
Query: 616 EDLIEYRGIREFKPKYQIKVDRFFFIPQEVHILSESSSIM---VRNNSIIGVNTPITLNK 672
EDL+E G++E + K Q K D + + + ES+ V + +N P TL K
Sbjct: 321 EDLLEQGGLKEVEKKNQRKADLLY------NAIEESNGFFRCPVEKSVRSLMNVPFTLEK 374
Query: 673 KSRVGGLVRVEKNKKKIELK 692
++ +K ++LK
Sbjct: 375 SELEAEFIKEAAKEKMVQLK 394
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,254,964
Number of Sequences: 26719
Number of extensions: 1281333
Number of successful extensions: 3009
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 2983
Number of HSP's gapped (non-prelim): 22
length of query: 1332
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1221
effective length of database: 8,352,787
effective search space: 10198752927
effective search space used: 10198752927
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 66 (30.0 bits)
Description of BAB33196.1

