
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
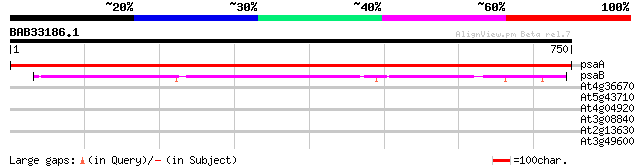
Query= BAB33186.1 750 aa
(750 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
psaA -chloroplast genome- PSI P700 apoprotein A1 1534 0.0
psaB -chloroplast genome- PSI P700 apoprotein A2 629 e-180
At4g36670 sugar transporter like protein 32 1.7
At5g43710 unknown protein 31 2.2
At4g04920 unknown protein 30 3.8
At3g08840 unknown protein 30 3.8
At2g13630 hypothetical protein 30 5.0
At3g49600 putative protein 30 6.5
>psaA -chloroplast genome- PSI P700 apoprotein A1
Length = 750
Score = 1534 bits (3971), Expect = 0.0
Identities = 732/750 (97%), Positives = 742/750 (98%)
Query: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT
Sbjct: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
Query: 61 SDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVG 120
SDLE+ISRK+FSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHI PSAQVVWPIVG
Sbjct: 61 SDLEEISRKVFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIGPSAQVVWPIVG 120
Query: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHK 180
QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIG LVFAALMLFAGWFHYHK
Sbjct: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGALVFAALMLFAGWFHYHK 180
Query: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF 240
AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF
Sbjct: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF 240
Query: 241 ILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
ILNRDLLAQLYPSFAEGATPFFTLNW+KY++FLTFRGGLDPVTGGLWLTDIAHHHLAIAI
Sbjct: 241 ILNRDLLAQLYPSFAEGATPFFTLNWSKYSEFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
Query: 301 LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSL 360
LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLS+NLAMLGSL
Sbjct: 301 LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSLNLAMLGSL 360
Query: 361 TIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
TIIVAHHMYSMPPYPYLATDY TQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPT RY
Sbjct: 361 TIIVAHHMYSMPPYPYLATDYATQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTNRY 420
Query: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 480
NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+F
Sbjct: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVF 480
Query: 481 AQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT 540
AQWIQNTHALAPG+TAPG T TSLTWGGG+LVAVG KVALLPIPLGTADFLVHHIHAFT
Sbjct: 481 AQWIQNTHALAPGVTAPGETASTSLTWGGGELVAVGGKVALLPIPLGTADFLVHHIHAFT 540
Query: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA
Sbjct: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
Query: 601 ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY
Sbjct: 601 ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
Query: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIV 720
GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSI+
Sbjct: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSII 720
Query: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
QGRAVGVTHYLLGGIATTWAFFLARIIAVG
Sbjct: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
>psaB -chloroplast genome- PSI P700 apoprotein A2
Length = 734
Score = 629 bits (1623), Expect = e-180
Identities = 344/746 (46%), Positives = 448/746 (59%), Gaps = 59/746 (7%)
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + + IF++HFGQL+IIFLW SG F
Sbjct: 8 FSQGLAQDP-TTRRIWFGIATAHDFESHDDITEERLYQNIFASHFGQLAIIFLWTSGNLF 66
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+E W+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 67 HVAWQGNFETWVQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGALGPVNIAYSGVYQWWYT 126
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L +AL L GW H K P+++WF++ ES LNHHL+GL G+
Sbjct: 127 IGLRTNEDLYTGALFLLFLSALSLIGGWLHLQPKWKPRVSWFKNAESRLNHHLSGLFGVS 186
Query: 207 SLSWAGHQVHVSLPI--------NQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFAE 256
SL+W GH VHV++P N FLN LPH L Q LY +
Sbjct: 187 SLAWTGHLVHVAIPASRGEYVRWNNFLNV--------LPHPQGLGPLFTGQWNLYAQNPD 238
Query: 257 GATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIG 316
++ F + LT GG P T LWLTD+AHHHLAIAILFLIAGHMYRTN+GIG
Sbjct: 239 SSSHLFGTSQGSGTAILTLLGGFHPQTQSLWLTDMAHHHLAIAILFLIAGHMYRTNFGIG 298
Query: 317 HGIKDILEAH--KGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPY 374
H IKD+LEAH G G+GHKGLY+ + S H QL + LA LG +T +VA HMYS+P Y
Sbjct: 299 HSIKDLLEAHIPPGGRLGRGHKGLYDTINNSIHFQLGLALASLGVITSLVAQHMYSLPAY 358
Query: 375 PYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAI 434
++A D+ TQ +L+THH +I GF++ GA AH AIF +RDY+P +++L R+L H++AI
Sbjct: 359 AFIAQDFTTQAALYTHHQYIAGFIMTGAFAHGAIFFIRDYNPEQNEDNVLARMLDHKEAI 418
Query: 435 ISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTH------ 488
ISHL+W +FLGFH+ GLY+HND M A G P+ I ++PIFAQWIQ+ H
Sbjct: 419 ISHLSWASLFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPIFAQWIQSAHGKTSYG 473
Query: 489 --ALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
L + P G S+ W G L A+ L + +G DFLVHH A +H T L
Sbjct: 474 FDVLLSSTSGPAFNAGRSI-WLPGWLNAINENSNSLFLTIGPGDFLVHHAIALGLHTTTL 532
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
IL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N I V F
Sbjct: 533 ILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLNTIGWVTF 592
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYG----S 662
++ WK + G++S F +SS + GWLRD+LW +SQ+I Y +
Sbjct: 593 YWHWKHITLWQGNVS-----------QFNESSTYLMGWLRDYLWLNSSQLINGYNPFGMN 641
Query: 663 SLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----ATQPRALS 718
SLS + FL H VWA MFL S RGYWQELIE++ WAH + +A +P ALS
Sbjct: 642 SLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLAWAHERTPLANLIRWKDKPVALS 701
Query: 719 IVQGRAVGVTHYLLGGIATTWAFFLA 744
IVQ R VG+ H+ +G I T AF +A
Sbjct: 702 IVQARLVGLAHFSVGYIFTYAAFLIA 727
>At4g36670 sugar transporter like protein
Length = 493
Score = 31.6 bits (70), Expect = 1.7
Identities = 22/79 (27%), Positives = 33/79 (40%), Gaps = 2/79 (2%)
Query: 134 GIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQDVES 193
GI T+ F +L T++GG+V A ML G A KLAW +
Sbjct: 321 GIMKTTFIFTATLLLDKVGRRKLLLTSVGGMVIALTMLGFGLTMAQNAGGKLAWALVLSI 380
Query: 194 MLNHHLAGL--LGLGSLSW 210
+ + +GLG ++W
Sbjct: 381 VAAYSFVAFFSIGLGPITW 399
>At5g43710 unknown protein
Length = 624
Score = 31.2 bits (69), Expect = 2.2
Identities = 19/61 (31%), Positives = 28/61 (45%), Gaps = 14/61 (22%)
Query: 104 DPTHIR---PSAQVVWPIV--------GQEILNGDVGGGFRGIQITSGFFQIWRASGITS 152
DP ++ SA +VWP+ G ++L GDV R + FF +W+ G T
Sbjct: 298 DPWYVEVNMDSAAIVWPVFNSLQAFWPGLQVLAGDVDPAIR---THTAFFSVWKRYGFTP 354
Query: 153 E 153
E
Sbjct: 355 E 355
>At4g04920 unknown protein
Length = 1196
Score = 30.4 bits (67), Expect = 3.8
Identities = 15/39 (38%), Positives = 23/39 (58%)
Query: 61 SDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYE 99
SD+ ++R ++SAH G+++I FL F G FS E
Sbjct: 516 SDIRTLARIVYSAHGGEIAIAFLRGGVHIFSGPTFSPVE 554
>At3g08840 unknown protein
Length = 519
Score = 30.4 bits (67), Expect = 3.8
Identities = 21/76 (27%), Positives = 39/76 (50%), Gaps = 8/76 (10%)
Query: 288 LTDIAHHHL-AIAILFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSW 346
L+++A H + A+ I+F + +G GI+++LE+H PF G G + +
Sbjct: 153 LSELAEHLVSAVDIVFPVI----HGRFGEDGGIQELLESHNIPFVGTGSRECFRAFD--- 205
Query: 347 HAQLSINLAMLGSLTI 362
+ S+ L LG +T+
Sbjct: 206 KYEASLELKELGFMTV 221
>At2g13630 hypothetical protein
Length = 297
Score = 30.0 bits (66), Expect = 5.0
Identities = 22/83 (26%), Positives = 37/83 (44%), Gaps = 5/83 (6%)
Query: 151 TSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQDVESMLNHHLAGLLGLGSLSW 210
T ++ + C A GL+ ++ A + AA + +D+ H +G+LG+G +
Sbjct: 186 TGKITIMCHASNGLLDLWVLEDANKGEWSTAAAVIPSIEDLVGNREHVFSGILGIGEII- 244
Query: 211 AGHQVHVSLPINQFLNAGVDPKE 233
+ LP N F DPKE
Sbjct: 245 ----LVPLLPPNTFFFLCYDPKE 263
>At3g49600 putative protein
Length = 1672
Score = 29.6 bits (65), Expect = 6.5
Identities = 13/35 (37%), Positives = 17/35 (48%)
Query: 604 VIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSS 638
+I WK D WG + + G+ I GN QSS
Sbjct: 1441 IIPELDWKYFCDEWGGLMENGISAFIEVGNTDQSS 1475
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.141 0.464
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,808,433
Number of Sequences: 26719
Number of extensions: 777560
Number of successful extensions: 1868
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1853
Number of HSP's gapped (non-prelim): 9
length of query: 750
length of database: 11,318,596
effective HSP length: 107
effective length of query: 643
effective length of database: 8,459,663
effective search space: 5439563309
effective search space used: 5439563309
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 64 (29.3 bits)
Description of BAB33186.1

