
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BAB33185.1 126 aa
(126 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
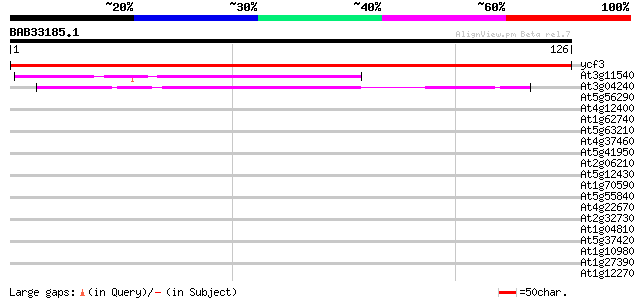
ycf3 -chloroplast genome- 253 2e-68
At3g11540 spindly (gibberellin signal transduction protein) 49 7e-07
At3g04240 O-linked GlcNAc transferase like protein 44 2e-05
At5g56290 peroxisomal targeting signal type 1 receptor 33 0.049
At4g12400 stress-induced protein sti1 -like protein 32 0.064
At1g62740 TPR-repeat protein 31 0.14
At5g63210 putative protein 31 0.19
At4g37460 unknown protein 31 0.19
At5g41950 unknown protein 30 0.24
At2g06210 putative TPR repeat nuclear phosphoprotein 30 0.32
At5g12430 putative protein 29 0.55
At1g70590 unknown protein 28 1.2
At5g55840 putative protein 28 1.6
At4g22670 HSP associated protein like 27 2.1
At2g32730 26S proteasome regulatory subunit 27 2.7
At1g04810 26S proteasome regulatory subunit, putative 27 2.7
At5g37420 contains similarity to unknown protein (pir||C71432) 27 3.5
At1g10980 membrane protein PTM1 precursor isolog 27 3.5
At1g27390 putative protein import receptor 26 4.6
At1g12270 putative protein 26 4.6
>ycf3 -chloroplast genome-
Length = 126
Score = 253 bits (645), Expect = 2e-68
Identities = 119/126 (94%), Positives = 123/126 (97%)
Query: 1 MSAQSEGNYAEALQNYYEAMRLEVDPYDRSYILYNIGLIHTSNGEHTKALEYYFRALERN 60
MSAQSEGNYAEALQNYYEAMRLE+DPYDRSYILYNIGLIHTSNGEHTKALEYYFRALERN
Sbjct: 1 MSAQSEGNYAEALQNYYEAMRLEIDPYDRSYILYNIGLIHTSNGEHTKALEYYFRALERN 60
Query: 61 PFLPQAFNNMAVICHYRGEQAIRQGDSEIAESWFDQAAEYWKQAIALTPGNYIEAQNWLK 120
PFLPQAFNNMAVICHYRGEQAI+QGDSE+AE+WF QAAEYWKQAI LTPGNYIEAQNWL
Sbjct: 61 PFLPQAFNNMAVICHYRGEQAIQQGDSEMAEAWFAQAAEYWKQAITLTPGNYIEAQNWLT 120
Query: 121 ITRRFE 126
ITRRFE
Sbjct: 121 ITRRFE 126
>At3g11540 spindly (gibberellin signal transduction protein)
Length = 914
Score = 48.9 bits (115), Expect = 7e-07
Identities = 29/79 (36%), Positives = 45/79 (56%), Gaps = 5/79 (6%)
Query: 2 SAQSEGNYAEALQNYYEAMRLEVDP-YDRSYILYNIGLIHTSNGEHTKALEYYFRALERN 60
S + GN E +Q YYEA L++DP Y +Y YN+G++++ ++ AL Y +A
Sbjct: 161 SLKLAGNTQEGIQKYYEA--LKIDPHYAPAY--YNLGVVYSEMMQYDNALSCYEKAALER 216
Query: 61 PFLPQAFNNMAVICHYRGE 79
P +A+ NM VI RG+
Sbjct: 217 PMYAEAYCNMGVIYKNRGD 235
Score = 33.9 bits (76), Expect = 0.022
Identities = 34/132 (25%), Positives = 54/132 (40%), Gaps = 31/132 (23%)
Query: 8 NYAEALQNYYEAMRLEVDPYDRSYILY---------------NIGLIHTSNGEHTKALEY 52
+YA+A+ N A E+ +D + + Y N+G+++ KA+E
Sbjct: 293 HYADAMYNLGVAYG-EMLKFDMAIVFYELAFHFNPHCAEACNNLGVLYKDRDNLDKAVEC 351
Query: 53 YFRALERNPFLPQAFNNMAVICHYRGEQAIRQGDSEIAESWFDQAAEYWKQAIALTPGNY 112
Y AL P Q+ NN+ V+ +G+ D AA ++AI P Y
Sbjct: 352 YQMALSIKPNFAQSLNNLGVVYTVQGK--------------MDAAASMIEKAILANP-TY 396
Query: 113 IEAQNWLKITRR 124
EA N L + R
Sbjct: 397 AEAFNNLGVLYR 408
>At3g04240 O-linked GlcNAc transferase like protein
Length = 977
Score = 44.3 bits (103), Expect = 2e-05
Identities = 33/111 (29%), Positives = 57/111 (50%), Gaps = 18/111 (16%)
Query: 7 GNYAEALQNYYEAMRLEVDPYDRSYILYNIGLIHTSNGEHTKALEYYFRALERNPFLPQA 66
G+ ALQ Y EA++L+ + +Y+ N+G ++ + G T+A+ Y AL+ P A
Sbjct: 237 GDLNRALQYYKEAVKLK-PAFPDAYL--NLGNVYKALGRPTEAIMCYQHALQMRPNSAMA 293
Query: 67 FNNMAVICHYRGEQAIRQGDSEIAESWFDQAAEYWKQAIALTPGNYIEAQN 117
F N+A I + +G+ D A ++KQA++ P ++EA N
Sbjct: 294 FGNIASIYYEQGQ--------------LDLAIRHYKQALSRDP-RFLEAYN 329
Score = 32.7 bits (73), Expect = 0.049
Identities = 25/105 (23%), Positives = 46/105 (43%), Gaps = 17/105 (16%)
Query: 7 GNYAEALQNYYEAMRLEVDPYDRSYILYNIGLIHTSNGEHTKALEYYFRALERNPFLPQA 66
G EA+ Y A+++ + + NI I+ G+ A+ +Y +AL R+P +A
Sbjct: 271 GRPTEAIMCYQHALQMRPNS---AMAFGNIASIYYEQGQLDLAIRHYKQALSRDPRFLEA 327
Query: 67 FNNMAVICHYRGEQAIRQGDSEIAESWFDQAAEYWKQAIALTPGN 111
+NN+ G++ D+A + Q +AL P +
Sbjct: 328 YNNL--------------GNALKDIGRVDEAVRCYNQCLALQPNH 358
Score = 26.9 bits (58), Expect = 2.7
Identities = 20/82 (24%), Positives = 35/82 (42%), Gaps = 23/82 (28%)
Query: 44 GEHTKALEYYFRALERNPFLPQAFNNMAVICHYRGEQAIRQGDSEIAESWFDQAAEYWKQ 103
G+ +A+ YY A+E P A++N+A +G +A + +Q
Sbjct: 135 GDTDRAIRYYLIAIELRPNFADAWSNLASAYMRKGR--------------LSEATQCCQQ 180
Query: 104 AIALTP---------GNYIEAQ 116
A++L P GN ++AQ
Sbjct: 181 ALSLNPLLVDAHSNLGNLMKAQ 202
>At5g56290 peroxisomal targeting signal type 1 receptor
Length = 728
Score = 32.7 bits (73), Expect = 0.049
Identities = 17/52 (32%), Positives = 33/52 (62%), Gaps = 3/52 (5%)
Query: 10 AEALQNYYEAMRLEVDPYDRSYILYNIGLIHTSNGEHTKALEYYFRALERNP 61
A+A+ Y +A+ L+ + Y R++ N+G+ + + G + +++ YY RAL NP
Sbjct: 641 ADAISAYQQALDLKPN-YVRAWA--NMGISYANQGMYKESIPYYVRALAMNP 689
>At4g12400 stress-induced protein sti1 -like protein
Length = 558
Score = 32.3 bits (72), Expect = 0.064
Identities = 28/111 (25%), Positives = 51/111 (45%), Gaps = 19/111 (17%)
Query: 2 SAQSEGNYAEALQNYYEAMRLEVDPYDRSYILY-NIGLIHTSNGEHTKALEYYFRALERN 60
+A S G+YA A+ ++ EA+ L ++ILY N + S + +AL + +E
Sbjct: 11 AAFSSGDYATAITHFTEAINLS----PTNHILYSNRSASYASLHRYEEALSDAKKTIELK 66
Query: 61 PFLPQAFNNMAVICHYRGEQAIRQGDSEIAESWFDQAAEYWKQAIALTPGN 111
P + ++ R G + I S FD+A + +K+ + + P N
Sbjct: 67 PDWSKGYS--------------RLGAAFIGLSKFDEAVDSYKKGLEIDPSN 103
>At1g62740 TPR-repeat protein
Length = 571
Score = 31.2 bits (69), Expect = 0.14
Identities = 22/76 (28%), Positives = 36/76 (46%), Gaps = 7/76 (9%)
Query: 3 AQSEGNYAEALQNYYEAMRLEVDPYDRSYILYNIGLIHTSNG-------EHTKALEYYFR 55
A +E E L+ EA R + + + Y NIG G ++ A+ +Y
Sbjct: 348 ALTEHRNPETLKRLNEAERAKKELEQQEYYDPNIGDEEREKGNDFFKEQKYPDAVRHYTE 407
Query: 56 ALERNPFLPQAFNNMA 71
A++RNP P+A++N A
Sbjct: 408 AIKRNPKDPRAYSNRA 423
Score = 26.9 bits (58), Expect = 2.7
Identities = 22/111 (19%), Positives = 49/111 (43%), Gaps = 19/111 (17%)
Query: 2 SAQSEGNYAEALQNYYEAMRLEVDPYDRSYILY-NIGLIHTSNGEHTKALEYYFRALERN 60
+A S G++ A+ ++ +A+ L +++L+ N H S + +AL + +E
Sbjct: 11 AAFSSGDFNSAVNHFTDAINLT----PTNHVLFSNRSAAHASLNHYDEALSDAKKTVELK 66
Query: 61 PFLPQAFNNMAVICHYRGEQAIRQGDSEIAESWFDQAAEYWKQAIALTPGN 111
P + ++ R G + + + FD+A E + + + + P N
Sbjct: 67 PDWGKGYS--------------RLGAAHLGLNQFDEAVEAYSKGLEIDPSN 103
Score = 25.4 bits (54), Expect = 7.9
Identities = 11/39 (28%), Positives = 22/39 (56%)
Query: 79 EQAIRQGDSEIAESWFDQAAEYWKQAIALTPGNYIEAQN 117
++A +G++ + F+ A ++ AI LTP N++ N
Sbjct: 3 DEAKAKGNAAFSSGDFNSAVNHFTDAINLTPTNHVLFSN 41
>At5g63210 putative protein
Length = 262
Score = 30.8 bits (68), Expect = 0.19
Identities = 20/74 (27%), Positives = 39/74 (52%), Gaps = 3/74 (4%)
Query: 1 MSAQSEGNYAEALQNYYEAMRLEVDPYDRSY-ILYNIGLIHTSNGEHTKALEYYFRALER 59
+S Q ++EA + Y A L V D+++ IL N+G ++ ++ + + +ALE
Sbjct: 138 ISLQLSDEHSEAEEVYKRA--LTVSKEDQAHAILSNLGNLYRQKKQYEVSKAMFSKALEL 195
Query: 60 NPFLPQAFNNMAVI 73
P A+NN+ ++
Sbjct: 196 KPGYAPAYNNLGLV 209
>At4g37460 unknown protein
Length = 869
Score = 30.8 bits (68), Expect = 0.19
Identities = 23/82 (28%), Positives = 35/82 (42%), Gaps = 4/82 (4%)
Query: 37 GLIHTSNGEHTKALEYYFRALERNPFLPQAFNNMAVICHYRGEQAIRQGDSEIAESWFDQ 96
G+ + G +TKA+ + + L+ P P+A ++ E D A
Sbjct: 304 GIAQVNEGNYTKAISIFDKVLKEEPTYPEALIGRGTAYAFQRELESAIADFTKAIQSNPA 363
Query: 97 AAEYWK---QAIALTPGNYIEA 115
A+E WK QA A G Y+EA
Sbjct: 364 ASEAWKRRGQARAAL-GEYVEA 384
>At5g41950 unknown protein
Length = 565
Score = 30.4 bits (67), Expect = 0.24
Identities = 30/123 (24%), Positives = 53/123 (42%), Gaps = 20/123 (16%)
Query: 10 AEALQNYYEAMRLEVDPYDRSYILYNIGLIHTSNGEHT--------------KALEYYFR 55
A A + Y A+ D +D LYN LI + ++ +A + Y
Sbjct: 206 AFAARKYASAIERNPDDHDA---LYNWALILQESADNVSPDSVSPSKDDLLEEACKKYDE 262
Query: 56 ALERNPFLPQAFNNMAVICHYRGEQAIRQGDSEIAESWFDQAAEYWKQAIALTPGNYIEA 115
A P L A+ N A+ ++A +G ++ AE ++QAA+ +++A+ L +
Sbjct: 263 ATRLCPTLYDAYYNWAIAI---SDRAKIRGRTKEAEELWEQAADNYEKAVQLNWNSSQAL 319
Query: 116 QNW 118
NW
Sbjct: 320 NNW 322
>At2g06210 putative TPR repeat nuclear phosphoprotein
Length = 501
Score = 30.0 bits (66), Expect = 0.32
Identities = 19/46 (41%), Positives = 27/46 (58%), Gaps = 1/46 (2%)
Query: 22 LEVDPYDRSYILYNIGLIHTSNGEHTKALEYYFRALERNPFLPQAF 67
LEV P D L +G ++T G++ KALEY +A + +P QAF
Sbjct: 131 LEVYP-DNCETLKALGHLYTQLGQNEKALEYMRKATKLDPRDAQAF 175
Score = 25.4 bits (54), Expect = 7.9
Identities = 12/39 (30%), Positives = 19/39 (47%)
Query: 29 RSYILYNIGLIHTSNGEHTKALEYYFRALERNPFLPQAF 67
+S+ YN+ + S G+ KA YY A++ P F
Sbjct: 65 KSHSFYNLARSYHSKGDFEKAGMYYMAAIKETNNNPHEF 103
>At5g12430 putative protein
Length = 1187
Score = 29.3 bits (64), Expect = 0.55
Identities = 25/88 (28%), Positives = 36/88 (40%), Gaps = 15/88 (17%)
Query: 18 EAMRLEVDPYDRSYILYNIGLIHT-----------SNGEHTKALEYYFRAL----ERNPF 62
E +RL+V P I N+ L+ +G HT+A+E+Y AL E PF
Sbjct: 878 ELLRLKVLPSSSMSIALNLHLLFRIQLPAAGNEAFQSGRHTEAVEHYTAALACNVESRPF 937
Query: 63 LPQAFNNMAVICHYRGEQAIRQGDSEIA 90
F N A G+ + D +A
Sbjct: 938 TAVCFCNRAAAYKALGQFSDAIADCSLA 965
>At1g70590 unknown protein
Length = 351
Score = 28.1 bits (61), Expect = 1.2
Identities = 24/97 (24%), Positives = 41/97 (41%), Gaps = 18/97 (18%)
Query: 33 LYNIGLIHTSNGEHTKALEYYFRALERNPFLPQAFNNMAVICHYRGEQAIRQGDSEIAES 92
+ + GL++ GE KA+ Y RA E + Q +A + ++ + + A
Sbjct: 145 MVDAGLVYWERGEKEKAVNLYRRASELGDAVGQCNLGIAYL-------QVQPSNPKEAMK 197
Query: 93 WFDQAAE------YWKQAIALTPG-----NYIEAQNW 118
W Q+AE ++ A+ L G N +EA W
Sbjct: 198 WLKQSAENGYVRAQYQLALCLHHGRVVQTNLLEATKW 234
>At5g55840 putative protein
Length = 1274
Score = 27.7 bits (60), Expect = 1.6
Identities = 20/56 (35%), Positives = 29/56 (51%), Gaps = 3/56 (5%)
Query: 5 SEGNYAEALQNYYEAMRLEVDPYDRSYILYNIGLIHTSNGEHTKALEYYFRALERN 60
SEGN+ EAL+ +Y + P + SY + GL N E A +Y R ++RN
Sbjct: 345 SEGNFKEALKMFYMMEAKGLTPSEVSYGVLLDGL--CKNAEFDLARGFYMR-MKRN 397
>At4g22670 HSP associated protein like
Length = 441
Score = 27.3 bits (59), Expect = 2.1
Identities = 25/93 (26%), Positives = 41/93 (43%), Gaps = 5/93 (5%)
Query: 1 MSAQSEGNYAEALQNYYEAMRLEVDPYDRSYILY-NIGLIHTSNGEHTKALEYYFRALER 59
M A SEGN+ EA+++ A+ L S I+Y N ++ + A+ ALE
Sbjct: 131 MEALSEGNFDEAIEHLTRAITLN----PTSAIMYGNRASVYIKLKKPNAAIRDANAALEI 186
Query: 60 NPFLPQAFNNMAVICHYRGEQAIRQGDSEIAES 92
NP + + + + GE A D +A +
Sbjct: 187 NPDSAKGYKSRGMARAMLGEWAEAAKDLHLAST 219
>At2g32730 26S proteasome regulatory subunit
Length = 1004
Score = 26.9 bits (58), Expect = 2.7
Identities = 11/23 (47%), Positives = 14/23 (60%)
Query: 26 PYDRSYILYNIGLIHTSNGEHTK 48
PY LY +GLIH ++GE K
Sbjct: 447 PYSEGGALYALGLIHANHGEGIK 469
>At1g04810 26S proteasome regulatory subunit, putative
Length = 1001
Score = 26.9 bits (58), Expect = 2.7
Identities = 11/23 (47%), Positives = 14/23 (60%)
Query: 26 PYDRSYILYNIGLIHTSNGEHTK 48
PY LY +GLIH ++GE K
Sbjct: 447 PYSEGGALYALGLIHANHGEGIK 469
>At5g37420 contains similarity to unknown protein (pir||C71432)
Length = 579
Score = 26.6 bits (57), Expect = 3.5
Identities = 14/34 (41%), Positives = 17/34 (49%)
Query: 20 MRLEVDPYDRSYILYNIGLIHTSNGEHTKALEYY 53
MR + Y RS ILYNI + H + K L Y
Sbjct: 33 MRQDSQQYQRSRILYNIWVSHNPSSLTLKLLMVY 66
>At1g10980 membrane protein PTM1 precursor isolog
Length = 516
Score = 26.6 bits (57), Expect = 3.5
Identities = 9/15 (60%), Positives = 10/15 (66%)
Query: 93 WFDQAAEYWKQAIAL 107
WF Q A+YWK I L
Sbjct: 238 WFPQVAQYWKDGIQL 252
>At1g27390 putative protein import receptor
Length = 208
Score = 26.2 bits (56), Expect = 4.6
Identities = 12/26 (46%), Positives = 17/26 (65%)
Query: 86 DSEIAESWFDQAAEYWKQAIALTPGN 111
D E A+ FD+A EY+++A PGN
Sbjct: 95 DPEEAKEHFDKATEYFQRAENEDPGN 120
>At1g12270 putative protein
Length = 572
Score = 26.2 bits (56), Expect = 4.6
Identities = 12/39 (30%), Positives = 22/39 (55%)
Query: 79 EQAIRQGDSEIAESWFDQAAEYWKQAIALTPGNYIEAQN 117
E+A +G++ + F A ++ +AIAL P N++ N
Sbjct: 3 EEAKAKGNAAFSSGDFTTAINHFTEAIALAPTNHVLFSN 41
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.133 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,853,336
Number of Sequences: 26719
Number of extensions: 108085
Number of successful extensions: 320
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 280
Number of HSP's gapped (non-prelim): 46
length of query: 126
length of database: 11,318,596
effective HSP length: 87
effective length of query: 39
effective length of database: 8,994,043
effective search space: 350767677
effective search space used: 350767677
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 54 (25.4 bits)
Description of BAB33185.1

