
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BAB33183.1 158 aa
(158 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
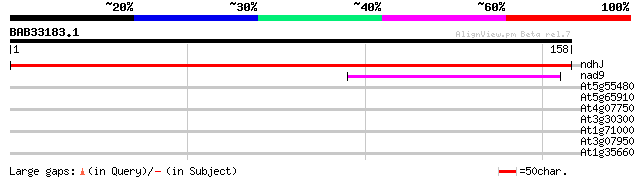
Score E
Sequences producing significant alignments: (bits) Value
ndhJ -chloroplast genome- NADH dehydrogenase subunit 303 3e-83
nad9 -mitochondrial genome- NADH dehydrogenase subunit 9 46 1e-05
At5g55480 predicted GPI-anchored protein 36 0.008
At5g65910 unknown protein 30 0.42
At4g07750 putative transposon protein 28 1.6
At3g30300 auxin-independent growth promoter-like protein 28 2.7
At1g71000 27 3.5
At3g07950 unknown protein 27 4.6
At1g35660 unknown protein 26 7.8
>ndhJ -chloroplast genome- NADH dehydrogenase subunit
Length = 158
Score = 303 bits (776), Expect = 3e-83
Identities = 139/158 (87%), Positives = 147/158 (92%)
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
MQG LS WL K GLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDV+
Sbjct: 1 MQGTLSVWLAKRGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVA 60
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGI 120
PGGLLASVYHLTRIE G+ Q EEVCIK+F R+NPRIPS+FWVWKS DFQERESYDMLGI
Sbjct: 61 PGGLLASVYHLTRIEYGVNQAEEVCIKVFTHRSNPRIPSVFWVWKSTDFQERESYDMLGI 120
Query: 121 SYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
+YD+HPRLKRILMPESWIGWPLRKDYI PNFYEIQDA+
Sbjct: 121 TYDSHPRLKRILMPESWIGWPLRKDYIAPNFYEIQDAY 158
>nad9 -mitochondrial genome- NADH dehydrogenase subunit 9
Length = 190
Score = 45.8 bits (107), Expect = 1e-05
Identities = 23/60 (38%), Positives = 35/60 (58%)
Query: 96 RIPSIFWVWKSADFQERESYDMLGISYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQ 155
RI + ++ SA ERE +DM G+S+ NHP L+RI + G PLRKD + +++
Sbjct: 97 RISPVVSLFPSAGRWEREVWDMFGVSFINHPDLRRISTDYGFEGHPLRKDLPLSGYVQVR 156
>At5g55480 predicted GPI-anchored protein
Length = 766
Score = 36.2 bits (82), Expect = 0.008
Identities = 42/166 (25%), Positives = 71/166 (42%), Gaps = 23/166 (13%)
Query: 5 LSAWLVKHGLVHRSLGFDYQ--GIETL-----QIKPED-WHSIAVILYVYGYNYLRSQCA 56
L+ ++K G++ RS FD GI T+ Q+KPE W ++ + +N S
Sbjct: 140 LTKVILKQGILSRSAAFDGNSYGISTVKDISTQLKPEGFWLNVQHDAFYAQHNLSMSSFL 199
Query: 57 YDVSPGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYD 116
+S ++ +L+ E ++ I RN P+ F + ++Y
Sbjct: 200 LSISKTVIID---YLSSPEVNFFRN----IGRRFGRNGPKFVFRFLEKDDVEVSTNQTYG 252
Query: 117 MLGISYDNHPRLKR----ILMPESWIGWPLRKDYITPNFYEIQDAH 158
L N LK +L+P+S+I WP+ Y+ P +QDAH
Sbjct: 253 SLA---GNLTFLKTFASGVLVPKSYI-WPIESQYLLPRTSFVQDAH 294
>At5g65910 unknown protein
Length = 432
Score = 30.4 bits (67), Expect = 0.42
Identities = 19/63 (30%), Positives = 36/63 (56%), Gaps = 2/63 (3%)
Query: 96 RIPSIFWVWKSADFQERESYDMLGISYDNHPRLKRILM-PESWIGWPLRKDYITPNFYEI 154
+I S F S D ++ YD++G++ + +K + M PE+W+ +PL D + + +E+
Sbjct: 122 KIASNFLQLSSEDVDPKD-YDVIGVTEELVAFVKDLAMHPETWLDFPLPDDDDSFDDFEL 180
Query: 155 QDA 157
DA
Sbjct: 181 ADA 183
>At4g07750 putative transposon protein
Length = 569
Score = 28.5 bits (62), Expect = 1.6
Identities = 18/52 (34%), Positives = 27/52 (51%), Gaps = 5/52 (9%)
Query: 97 IPSIFWVWKSAD-FQERESYDMLGISYDNHPRL--KRILMPESWIGWPLRKD 145
I ++ WKS D R+ ++LGI D HP+ KR +P + W L K+
Sbjct: 509 ISTLLHCWKSKDGLNARKDLEVLGIRKDLHPKTKGKRTYLPAAL--WSLSKE 558
>At3g30300 auxin-independent growth promoter-like protein
Length = 677
Score = 27.7 bits (60), Expect = 2.7
Identities = 12/44 (27%), Positives = 21/44 (47%)
Query: 7 AWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNY 50
AW++K G+V L D + P + ++L YGY++
Sbjct: 309 AWMIKRGIVKGKLSVDSAEQRLAGLCPLMPEEVGILLRAYGYSW 352
>At1g71000
Length = 146
Score = 27.3 bits (59), Expect = 3.5
Identities = 14/52 (26%), Positives = 26/52 (49%), Gaps = 8/52 (15%)
Query: 115 YDMLGISYDN--------HPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
Y++LG++ D+ + +L +I P+ W P R F +IQ+A+
Sbjct: 10 YEILGVAVDSSAEQIRRAYHKLAKIWHPDRWTKDPFRSGEAKRRFQQIQEAY 61
>At3g07950 unknown protein
Length = 304
Score = 26.9 bits (58), Expect = 4.6
Identities = 11/46 (23%), Positives = 24/46 (51%), Gaps = 5/46 (10%)
Query: 36 WHSIAVILYVYGYNYLRSQCAYDVSPGGLLASVYHLTRIECGIYQP 81
W S + +++ N+L C + + ++Y++TR+E +Y P
Sbjct: 85 WGSTEFLKFIFVVNFLTYLCVFVTA-----IALYYITRLEVYLYMP 125
>At1g35660 unknown protein
Length = 1405
Score = 26.2 bits (56), Expect = 7.8
Identities = 14/48 (29%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Query: 78 IYQPEEVCIKIFVPRNNPRIPSIFWVWKSADFQERESYDMLGISYDNH 125
+ Q + + F+ N P +W++KSA E + +D L I NH
Sbjct: 416 VVQQNGLAVLRFLQSNCKEDPGAYWLYKSAGEDELQLFD-LSIISKNH 462
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.143 0.474
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,137,383
Number of Sequences: 26719
Number of extensions: 176565
Number of successful extensions: 243
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 241
Number of HSP's gapped (non-prelim): 9
length of query: 158
length of database: 11,318,596
effective HSP length: 91
effective length of query: 67
effective length of database: 8,887,167
effective search space: 595440189
effective search space used: 595440189
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 56 (26.2 bits)
Description of BAB33183.1

