
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
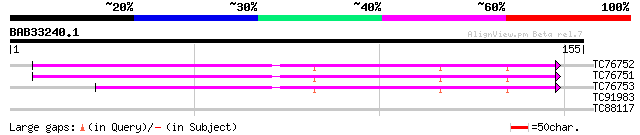
Query= BAB33240.1 155 aa
(155 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76752 homologue to SP|O65731|RS5_CICAR 40S ribosomal protein S... 63 5e-11
TC76751 similar to GP|15294021|gb|AAK95187.1 40S ribosomal prote... 63 5e-11
TC76753 SP|O65731|RS5_CICAR 40S ribosomal protein S5 (Fragment).... 59 1e-09
TC91983 weakly similar to GP|18376255|emb|CAD21369. probable GEL... 30 0.39
TC88117 weakly similar to PIR|E96803|E96803 probable lipase 416... 25 9.6
>TC76752 homologue to SP|O65731|RS5_CICAR 40S ribosomal protein S5
(Fragment). [Chickpea Garbanzo] {Cicer arietinum},
partial (97%)
Length = 943
Score = 62.8 bits (151), Expect = 5e-11
Identities = 51/150 (34%), Positives = 80/150 (53%), Gaps = 7/150 (4%)
Frame = +2
Query: 7 AEKKTAKSDPIYRNRLVNMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLSVLRQAI 66
++K+ K+ RL N L+ +GKK +A +II AM+ I T+ NP+ V+ A+
Sbjct: 266 SKKRFRKAQCPIVERLTNSLMMHGRNNGKKLMAVRIIKHAMEIIHLLTDQNPIQVIVDAV 445
Query: 67 RGVTPDIAVKARRVG--GSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRN---MAFKLS 121
P A R+G G + ++I + AI L +R+ RN +A L+
Sbjct: 446 VNSGP--REDATRIGSAGVVRRQAVDISPLRRVNQAIYLLTTGAREAAFRNIKTIAECLA 619
Query: 122 SELVDAAKGSGD--AIRKKEETHRMAEANR 149
EL++AAKGS + AI+KK+E R+A+ANR
Sbjct: 620 DELINAAKGSSNSYAIKKKDEIERVAKANR 709
>TC76751 similar to GP|15294021|gb|AAK95187.1 40S ribosomal protein S5
{Ictalurus punctatus}, partial (93%)
Length = 910
Score = 62.8 bits (151), Expect = 5e-11
Identities = 51/150 (34%), Positives = 80/150 (53%), Gaps = 7/150 (4%)
Frame = +1
Query: 7 AEKKTAKSDPIYRNRLVNMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLSVLRQAI 66
++K+ K+ RL N L+ +GKK +A +II AM+ I T+ NP+ V+ A+
Sbjct: 250 SKKRFRKAQCPIVERLTNSLMMHGRNNGKKLMAVRIIKHAMEIIHLLTDQNPIQVIVDAV 429
Query: 67 RGVTPDIAVKARRVG--GSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRN---MAFKLS 121
P A R+G G + ++I + AI L +R+ RN +A L+
Sbjct: 430 VNSGP--REDATRIGSAGVVRRQAVDISPLRRVNQAIYLLTTGAREAAFRNIKTIAECLA 603
Query: 122 SELVDAAKGSGD--AIRKKEETHRMAEANR 149
EL++AAKGS + AI+KK+E R+A+ANR
Sbjct: 604 DELINAAKGSSNSYAIKKKDEIERVAKANR 693
>TC76753 SP|O65731|RS5_CICAR 40S ribosomal protein S5 (Fragment). [Chickpea
Garbanzo] {Cicer arietinum}, partial (66%)
Length = 1221
Score = 58.5 bits (140), Expect = 1e-09
Identities = 47/133 (35%), Positives = 72/133 (53%), Gaps = 7/133 (5%)
Frame = +3
Query: 24 NMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLSVLRQAIRGVTPDIAVKARRVG-- 81
N L+ +GKK +A +II AM+ I T+ NP+ V+ A+ P A R+G
Sbjct: 666 NSLMMHGRNNGKKLMAVRIIKHAMEIIHLLTDQNPIQVIVDAVVNSGP--REDATRIGSA 839
Query: 82 GSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRN---MAFKLSSELVDAAKGSGD--AIR 136
G + ++I + AI L +R+ RN +A L+ EL++AAKGS + AI+
Sbjct: 840 GVVRRQAVDISPLRRVNQAIYLLTTGAREAAFRNIKTIAECLADELINAAKGSSNSYAIK 1019
Query: 137 KKEETHRMAEANR 149
KK+E R+A+ANR
Sbjct: 1020KKDEIERVAKANR 1058
>TC91983 weakly similar to GP|18376255|emb|CAD21369. probable GEL1 protein
{Neurospora crassa}, partial (38%)
Length = 973
Score = 30.0 bits (66), Expect = 0.39
Identities = 18/68 (26%), Positives = 30/68 (43%), Gaps = 3/68 (4%)
Frame = -1
Query: 68 GVTPDIAVKARRVGGSTHQVPIEIGS---TQGKALAIRWLLGASRKRPGRNMAFKLSSEL 124
GVTP + V+ + V +E+GS T G ++ R K GR++ F ++ +
Sbjct: 316 GVTPGVVVEGKEVAAGIIATTVEVGSSLHTVGSNVSSRVTNWHVTKAAGRHVVFDVTGDS 137
Query: 125 VDAAKGSG 132
D G
Sbjct: 136 FDVGSRQG 113
>TC88117 weakly similar to PIR|E96803|E96803 probable lipase 4162-5963
[imported] - Arabidopsis thaliana, partial (35%)
Length = 775
Score = 25.4 bits (54), Expect = 9.6
Identities = 12/31 (38%), Positives = 18/31 (57%)
Frame = -1
Query: 81 GGSTHQVPIEIGSTQGKALAIRWLLGASRKR 111
GG+T + IG+T ++ I WLL + KR
Sbjct: 157 GGNTSSAILHIGATSITSIPIIWLLQINFKR 65
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.131 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,269,878
Number of Sequences: 36976
Number of extensions: 29351
Number of successful extensions: 125
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 125
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 125
length of query: 155
length of database: 9,014,727
effective HSP length: 88
effective length of query: 67
effective length of database: 5,760,839
effective search space: 385976213
effective search space used: 385976213
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 54 (25.4 bits)
Description of BAB33240.1

