
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BAB33231.1 134 aa
(134 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
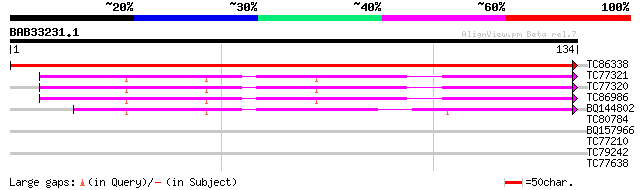
TC86338 similar to GP|13359014|dbj|BAB33231. ribosomal protein S... 238 7e-64
TC77321 homologue to GP|440824|gb|AAA61608.1|| ribosomal protein... 44 2e-05
TC77320 homologue to GP|440824|gb|AAA61608.1|| ribosomal protein... 44 2e-05
TC86986 GP|440824|gb|AAA61608.1|| ribosomal protein S15 {Arabido... 43 4e-05
BQ144802 similar to GP|13560779|gb cytoplasmic ribosomal protein... 40 4e-04
TC80784 similar to PIR|T50352|T50352 ornithine--oxo-acid transam... 30 0.28
BQ157966 27 2.4
TC77210 similar to GP|21954717|gb|AAM83095.1 SOS2-like protein k... 27 3.1
TC79242 25 9.1
TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protei... 25 9.1
>TC86338 similar to GP|13359014|dbj|BAB33231. ribosomal protein S8 {Lotus
japonicus}, complete
Length = 1441
Score = 238 bits (606), Expect = 7e-64
Identities = 118/134 (88%), Positives = 126/134 (93%)
Frame = +3
Query: 1 MGKDTIADIITSIRNADMNRKRTVQIPFTNITENTVKILLREGFIENVRKHRESDKSFLV 60
+GKDTIAD++T IRNADMNRKRTVQIP TNITEN VKILLREGF+ENVRKH ES+K FLV
Sbjct: 447 VGKDTIADMLTLIRNADMNRKRTVQIPLTNITENIVKILLREGFVENVRKHGESNKYFLV 626
Query: 61 LTLRYRRNTKGSYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREA 120
LTLRYRRN K SYKTFLNLKRISTPGLRIY NYQ+IPR+LGGMGIVILSTSRGIMTDREA
Sbjct: 627 LTLRYRRNRKESYKTFLNLKRISTPGLRIYSNYQQIPRVLGGMGIVILSTSRGIMTDREA 806
Query: 121 RLEKIGGEVLCYIW 134
RLE+IGGEVLCYIW
Sbjct: 807 RLERIGGEVLCYIW 848
>TC77321 homologue to GP|440824|gb|AAA61608.1|| ribosomal protein S15
{Arabidopsis thaliana}, complete
Length = 699
Score = 43.5 bits (101), Expect = 2e-05
Identities = 39/133 (29%), Positives = 64/133 (47%), Gaps = 6/133 (4%)
Frame = +3
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + S+ NA+ KR V I P + + + ++ + G+I E V HR +V+ L
Sbjct: 96 DALKSMYNAEKRGKRQVMIRPSSKVIIKFLMVMQKHGYIGEFEYVDDHRSGK---IVVEL 266
Query: 64 RYRRNTKG--SYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREAR 121
R N G S + + +K I R+ + Q G ++L+TS GIM EAR
Sbjct: 267 NGRLNKCGVISPRFDVGVKEIEGWTARLLPSRQ--------FGYIVLTTSAGIMDHEEAR 422
Query: 122 LEKIGGEVLCYIW 134
+ +GG+VL + +
Sbjct: 423 RKNVGGKVLGFFY 461
>TC77320 homologue to GP|440824|gb|AAA61608.1|| ribosomal protein S15
{Arabidopsis thaliana}, complete
Length = 649
Score = 43.5 bits (101), Expect = 2e-05
Identities = 39/133 (29%), Positives = 64/133 (47%), Gaps = 6/133 (4%)
Frame = +1
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + S+ NA+ KR V I P + + + ++ + G+I E V HR +V+ L
Sbjct: 85 DALKSMYNAEKRGKRQVMIRPSSKVIIKFLMVMQKHGYIGEFEYVDDHRSGK---IVVEL 255
Query: 64 RYRRNTKG--SYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREAR 121
R N G S + + +K I R+ + Q G ++L+TS GIM EAR
Sbjct: 256 NGRLNKCGVISPRFDVGVKEIEGWTARLLPSRQ--------FGYIVLTTSAGIMDHEEAR 411
Query: 122 LEKIGGEVLCYIW 134
+ +GG+VL + +
Sbjct: 412 RKNVGGKVLGFFY 450
>TC86986 GP|440824|gb|AAA61608.1|| ribosomal protein S15 {Arabidopsis
thaliana}, complete
Length = 728
Score = 42.7 bits (99), Expect = 4e-05
Identities = 39/133 (29%), Positives = 64/133 (47%), Gaps = 6/133 (4%)
Frame = +1
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + S+ NA+ KR V I P + + + ++ + G+I E V HR +V+ L
Sbjct: 130 DALKSMYNAEKRGKRQVMIRPSSKVIIKFLIVMQKHGYIGEFEYVDDHRSGK---IVVEL 300
Query: 64 RYRRNTKG--SYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREAR 121
R N G S + + +K I R+ + Q G ++L+TS GIM EAR
Sbjct: 301 NGRLNKCGVISPRFDVGVKEIEGWTARLLPSRQ--------FGYIVLTTSAGIMDHEEAR 456
Query: 122 LEKIGGEVLCYIW 134
+ +GG+VL + +
Sbjct: 457 RKNVGGKVLGFFY 495
>BQ144802 similar to GP|13560779|gb cytoplasmic ribosomal protein S15a
{Daucus carota}, partial (87%)
Length = 498
Score = 39.7 bits (91), Expect = 4e-04
Identities = 36/125 (28%), Positives = 59/125 (46%), Gaps = 6/125 (4%)
Frame = +1
Query: 16 ADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTLRYRRNTKG 71
A+ KR V I P + + ++++++ G+I E V HR +V+ L R N G
Sbjct: 79 AEKRGKRQVLIRPSSKVVIKFLQVMMKHGYIGEFEIVDDHRAGK---IVVELNGRLNKCG 249
Query: 72 SYKTFLNLKRISTPGLRIYYNYQKIPRILGG--MGIVILSTSRGIMTDREARLEKIGGEV 129
++K + R+L G +IL+TS GIM EAR +K+GG+V
Sbjct: 250 VISPRYDVKHTEIEAW--------VARLLPSRLFGKIILTTSSGIMDHEEARRKKVGGKV 405
Query: 130 LCYIW 134
L + +
Sbjct: 406 LGFFY 420
>TC80784 similar to PIR|T50352|T50352 ornithine--oxo-acid transaminase (EC
2.6.1.13) [imported] - fission yeast, partial (36%)
Length = 1206
Score = 30.0 bits (66), Expect = 0.28
Identities = 12/31 (38%), Positives = 21/31 (67%)
Frame = -1
Query: 104 GIVILSTSRGIMTDREARLEKIGGEVLCYIW 134
G ++L+TS GIM EAR + GG+++ + +
Sbjct: 1020 GYLVLTTSAGIMDHHEARRKHTGGKIIGFFY 928
>BQ157966
Length = 319
Score = 26.9 bits (58), Expect = 2.4
Identities = 13/29 (44%), Positives = 19/29 (64%)
Frame = +3
Query: 56 KSFLVLTLRYRRNTKGSYKTFLNLKRIST 84
KS L++ + Y+ T G Y F+N+KRI T
Sbjct: 51 KSSLLIFITYQAETLG-YNPFINIKRIKT 134
>TC77210 similar to GP|21954717|gb|AAM83095.1 SOS2-like protein kinase
{Glycine max}, partial (88%)
Length = 1831
Score = 26.6 bits (57), Expect = 3.1
Identities = 16/63 (25%), Positives = 30/63 (47%)
Frame = -2
Query: 14 RNADMNRKRTVQIPFTNITENTVKILLREGFIENVRKHRESDKSFLVLTLRYRRNTKGSY 73
+++ N K P NI+ +TV LLR+ N+R+ +L+ +R +T+
Sbjct: 948 KSSQKNVKNNTTRP--NISLSTVVTLLRDNLRRNIRRSTTCGMQQSILSQMFRESTETKI 775
Query: 74 KTF 76
+ F
Sbjct: 774 RDF 766
>TC79242
Length = 1802
Score = 25.0 bits (53), Expect = 9.1
Identities = 19/66 (28%), Positives = 31/66 (46%), Gaps = 5/66 (7%)
Frame = -1
Query: 6 IADIITSIRNADMNRKRTVQIPFTNITENTVKILLREGF-----IENVRKHRESDKSFLV 60
I I S + ++++K ++ + + N V LR GF IE R HR + K+F
Sbjct: 845 ITQITKSFHHLNLHQKLSLFLQAFPLNRNWVA--LRSGFPIRVFIETPRFHRSTQKNFHC 672
Query: 61 LTLRYR 66
L L +
Sbjct: 671 LNLHQK 654
>TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protein {Oryza
sativa (japonica cultivar-group)}, partial (62%)
Length = 2345
Score = 25.0 bits (53), Expect = 9.1
Identities = 11/49 (22%), Positives = 27/49 (54%)
Frame = +2
Query: 20 RKRTVQIPFTNITENTVKILLREGFIENVRKHRESDKSFLVLTLRYRRN 68
RK+ +++P IT+ + LLR + V++ ++ ++ ++ +RN
Sbjct: 665 RKQRMKVPIQKITKMAKRRLLRNLNLMRVKRKMNQRRTTSLIQMKVKRN 811
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.141 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,323,863
Number of Sequences: 36976
Number of extensions: 36309
Number of successful extensions: 135
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 135
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 135
length of query: 134
length of database: 9,014,727
effective HSP length: 86
effective length of query: 48
effective length of database: 5,834,791
effective search space: 280069968
effective search space used: 280069968
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 53 (25.0 bits)
Description of BAB33231.1

