
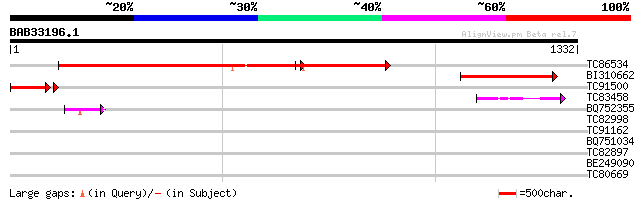
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BAB33196.1 1332 aa
(1332 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86534 homologue to SP|P12227|RPOD_PEA DNA-directed RNA polymer... 968 0.0
BI310662 homologue to SP|P12227|RPO DNA-directed RNA polymerase ... 400 e-111
TC91500 similar to SP|Q9BBS8|RPOC_LOTJA DNA-directed RNA polymer... 182 5e-48
TC83458 homologue to PIR|T07796|T07796 DNA-directed RNA polymera... 49 1e-05
BQ752355 similar to PIR|A00694|RN DNA-directed RNA polymerase (E... 47 6e-05
TC82998 weakly similar to PIR|A00694|RNBY3L DNA-directed RNA pol... 43 0.001
TC91162 similar to PIR|T06754|T06754 DNA-directed RNA polymerase... 42 0.002
BQ751034 similar to GP|18307453|e probable DNA-directed RNA poly... 39 0.017
TC82897 similar to GP|16604396|gb|AAL24204.1 AT5g18630/T1A4_10 {... 32 1.3
BE249090 30 4.8
TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 30 8.1
>TC86534 homologue to SP|P12227|RPOD_PEA DNA-directed RNA polymerase beta'
chain (EC 2.7.7.6) (Fragment). [Garden pea] {Pisum
sativum}, partial (61%)
Length = 2441
Score = 968 bits (2502), Expect = 0.0
Identities = 484/614 (78%), Positives = 531/614 (85%), Gaps = 34/614 (5%)
Frame = +1
Query: 115 MNPNFSMTDPFNPVHIMSFSGARGNASQVHQLVGMRGLMSDPQGQMIDLPIQSNLREGLS 174
+NPNF MTDPFNPVHIMSFSGARGNASQVHQLVGMRGLMSDPQGQMIDLPIQSNLREGLS
Sbjct: 7 VNPNFRMTDPFNPVHIMSFSGARGNASQVHQLVGMRGLMSDPQGQMIDLPIQSNLREGLS 186
Query: 175 LTEYIISCYGARKGVVDTAVRTSDAGYLTRRLVEVVQHIVVRRTDCGTVRGISVNTRNRM 234
LTEYIISCYGARKGVVDTAVRTSDAGYLTRRLVEVVQHIVVRRTDCGT+RGISVNTRN M
Sbjct: 187 LTEYIISCYGARKGVVDTAVRTSDAGYLTRRLVEVVQHIVVRRTDCGTIRGISVNTRNGM 366
Query: 235 MSERILIQTLIGRVLADDIYIGSRCIVVRNQDIGIGLINRFINFQTQPIFIRTPFTCRNT 294
+ ERILIQTLIGRV+ADDIYIGSRCIVVRNQDIGIGLINRFI FQTQPIFIRTPFTCRNT
Sbjct: 367 IPERILIQTLIGRVVADDIYIGSRCIVVRNQDIGIGLINRFITFQTQPIFIRTPFTCRNT 546
Query: 295 SWICRLCYGRSPIHGNLVELGEAVGIIAGQSIGEPGTQLTLRTFHTGGVFTGGTAEYVRS 354
SWICRLCYGRSPIHG+LVELGEAVGIIAGQSIGEPGTQLTLRTFHTGGVFTGGTAEY R+
Sbjct: 547 SWICRLCYGRSPIHGDLVELGEAVGIIAGQSIGEPGTQLTLRTFHTGGVFTGGTAEYARA 726
Query: 355 PSNGKIKFNENSAYPTRTRHGHPAFLCYIDLYVTIESNDIMHNVIIPPKSFLLVQNDQYV 414
PSNGKIKFNE+ +PTRTRHG+PAF+C IDLYVT+ES+DI+HNVIIPPKSFLLVQNDQYV
Sbjct: 727 PSNGKIKFNEDLVHPTRTRHGYPAFICNIDLYVTVESDDIIHNVIIPPKSFLLVQNDQYV 906
Query: 415 KSEQIIAEIRAGTYTLNLKEKVRKHIFSDSEGEMHWSTNIYHVSEFAYSNVHILPKTSHL 474
KSEQ+IAEIRAGT T NLKE+VRKHI+SDSEGEMHWST++YH SEF YSNVHILPKTSHL
Sbjct: 907 KSEQVIAEIRAGTSTFNLKERVRKHIYSDSEGEMHWSTDVYHASEFLYSNVHILPKTSHL 1086
Query: 475 WILSGNSHKSDTVSLSLLKDQDQMSTHSLPTAKRNTSNFLVSNNQVRL------------ 522
WILSG S +SDT+ L KDQDQ++ SL T KRN SN LVSN++V+L
Sbjct: 1087WILSGKSCRSDTIHFLLRKDQDQINIDSLSTGKRNISNLLVSNDEVKLKLLSLKTFGTKE 1266
Query: 523 -------------CPDHCHFMHPTISPDTSNLLAKKRRNRFIIPFLFRSIRERNNELMP- 568
C DH H M+P I DT LLAK+RRNRF+IP F+SI+ER NELM
Sbjct: 1267KGISNYSILNQMICTDHSHLMYPAIFHDTFYLLAKRRRNRFLIP--FQSIQERKNELMRP 1440
Query: 569 -DISVEIPIDGIIHKNSILAYFDDPQYRTQSSGIAKYKTIGIHSIFQKEDLIEYRGIREF 627
+S+EIPI+GI H+NSI AYFDDPQYR Q+SGI KY+T+GIHS+FQKED IEYRGI+E
Sbjct: 1441FGVSIEIPINGIFHRNSIFAYFDDPQYRRQNSGITKYRTVGIHSVFQKEDFIEYRGIKEL 1620
Query: 628 KPKYQIKVDRFFFIPQEVHILSESSSIMVRNNSIIGVNTPITLNKKSRVGGLVRVEKNKK 687
KPKYQIKVDRFFFIP+EVHIL ESSSIMVRNNS++G+ TPIT N +SRVGGLVR+EK KK
Sbjct: 1621KPKYQIKVDRFFFIPEEVHILPESSSIMVRNNSLVGIGTPITFNIRSRVGGLVRLEKKKK 1800
Query: 688 K-------IELKIF 694
K +E+ IF
Sbjct: 1801KKSN*NYFLEISIF 1842
Score = 328 bits (842), Expect = 6e-90
Identities = 173/225 (76%), Positives = 194/225 (85%), Gaps = 1/225 (0%)
Frame = +3
Query: 672 KKSRVGGLVRVEKNKKKIELKIFSGDIHFPGEIDKISQHSAILIPPEMVKKKNSKESKKK 731
K SR G +R EK KKKIELK+FSG+IHFPGE DKIS+HS+ILIPP VKKK KESKK
Sbjct: 1758 KSSRRIGPIRKEK-KKKIELKLFSGNIHFPGERDKISRHSSILIPPGTVKKKR-KESKKI 1931
Query: 732 TNWRYIQWITTTKKKYFVLVRPVILYDIADSINLVKLFPQDLFKEWDNLELKVLNFILYG 791
NW Y+QWI TTKKKYFVLVRPVILY+I DSI+ +KLFPQDLF+E DNLELKV+N+ILYG
Sbjct: 1932 NNWIYVQWIATTKKKYFVLVRPVILYEIPDSIDFIKLFPQDLFQERDNLELKVVNYILYG 2111
Query: 792 NGKSIRGILDTSIQLVRTCLVLNWNEDEKSSSIEEALASFVEVSTNGLIRYFLRIDLVKS 851
NGKSIRGI DT IQLVRTCLVLNW++ +KS SIEEA +SFVEVSTNGLI YFLRIDLVKS
Sbjct: 2112 NGKSIRGISDTRIQLVRTCLVLNWDQGKKSPSIEEAPSSFVEVSTNGLIEYFLRIDLVKS 2291
Query: 852 HISYIRKRNDPSSSGLISYNESDRININPFFSI-YKENIQQSLSQ 895
+ +YIRKRNDPS GLI+ NESDRININPFFSI YK I++SLSQ
Sbjct: 2292 NTAYIRKRNDPSGLGLIADNESDRININPFFSIHYKAKIKKSLSQ 2426
>BI310662 homologue to SP|P12227|RPO DNA-directed RNA polymerase beta' chain
(EC 2.7.7.6) (Fragment). [Garden pea] {Pisum sativum},
partial (19%)
Length = 688
Score = 400 bits (1029), Expect = e-111
Identities = 208/229 (90%), Positives = 217/229 (93%), Gaps = 2/229 (0%)
Frame = +2
Query: 1060 GPNLK--SGQVITVQMDFVGIRLANPYLATPGATIHGHYGEMLYEGDILVTFIYEKSRSG 1117
GP+LK SGQVITVQMD V IR ANPYLATP ATIHGHYGE+LY+GDILVTFIYEKSRS
Sbjct: 2 GPHLKLKSGQVITVQMDSVIIRSANPYLATPEATIHGHYGEILYQGDILVTFIYEKSRSS 181
Query: 1118 DITQGLPKVEQVLEVRSIDSISMNLEKRIDAWNERITGILGIPWRFLIGAELTIAQSRIS 1177
DITQGLPKVEQ+LE+RSIDSISMNLEKRIDAWNE IT ILGIPW FLIGAELTIAQSRIS
Sbjct: 182 DITQGLPKVEQILEIRSIDSISMNLEKRIDAWNECITRILGIPWGFLIGAELTIAQSRIS 361
Query: 1178 LVNKIQRVYRSQGVHIHNRHIEIIVRQITSKVLVSEDGMSNVFSPGELIGLLRAQRTGRA 1237
LVNKIQ+VYRSQGVHIHNRHIEIIVRQITSKVLVSEDGMSNVF PGELIGLLRA+RTGRA
Sbjct: 362 LVNKIQKVYRSQGVHIHNRHIEIIVRQITSKVLVSEDGMSNVFLPGELIGLLRAERTGRA 541
Query: 1238 LEESICYRTLLLGITKTSLNTQSFISEASFQETARVLAKAALRGRIDWL 1286
LEE+ICYR LLLGITKTSLNTQSFISEASFQETARVLAKAALRGRIDW+
Sbjct: 542 LEEAICYRALLLGITKTSLNTQSFISEASFQETARVLAKAALRGRIDWV 688
>TC91500 similar to SP|Q9BBS8|RPOC_LOTJA DNA-directed RNA polymerase beta'
chain (EC 2.7.7.6). {Lotus japonicus}, partial (22%)
Length = 1004
Score = 182 bits (462), Expect(2) = 5e-48
Identities = 90/95 (94%), Positives = 92/95 (96%)
Frame = +1
Query: 1 MAERANLVFHNKVIGGTAIKRIISRLIDHFGMAYTSHILDQVKTLGFRQATTTSISLGID 60
MAERANLVFHN VIGGTAIKR+ISRLIDHFGMAYTSHILDQVKTLGFRQAT TSISLGID
Sbjct: 661 MAERANLVFHNNVIGGTAIKRLISRLIDHFGMAYTSHILDQVKTLGFRQATATSISLGID 840
Query: 61 DLLTIPSKGWLVQDAEQQSLILEKHHHYGNVHAVE 95
DLLTIPSKGWLVQDAEQQS ILEKH+HYGNVHAVE
Sbjct: 841 DLLTIPSKGWLVQDAEQQSSILEKHNHYGNVHAVE 945
Score = 28.9 bits (63), Expect(2) = 5e-48
Identities = 12/13 (92%), Positives = 12/13 (92%)
Frame = +2
Query: 103 IWYATSEYLRQEM 115
IW ATSEYLRQEM
Sbjct: 965 IWXATSEYLRQEM 1003
>TC83458 homologue to PIR|T07796|T07796 DNA-directed RNA polymerase (EC
2.7.7.6) largest chain - soybean (fragment), partial
(63%)
Length = 1220
Score = 48.9 bits (115), Expect = 1e-05
Identities = 53/213 (24%), Positives = 91/213 (41%), Gaps = 6/213 (2%)
Frame = +1
Query: 1098 EMLYEG--DILVTFIYE-KSRSGDITQGL-PKVEQVLEVRSIDSISMNLEKRIDAWNERI 1153
EM G DI FI K + D +G P E +L+ ++ +++ + +DA +R
Sbjct: 70 EMTLRGIPDINKVFIKNTKIQKFDENEGFKPHEEWMLDTEGVNLLAVMCHEDVDA--KRT 243
Query: 1154 TGILGIPWRFLIGAELTIAQSRISLVNKIQRVYRSQGVHIHNRHIEIIVRQITSKVLVSE 1213
T I ++G E R SL+++++ V G +++ RH+ I+ +T
Sbjct: 244 TSNHLIEVIEVLGIEAV----RRSLLDELRVVISFDGSYVNYRHLAILCDTMT------- 390
Query: 1214 DGMSNVFSPGELIGLLRAQRTGRALEESICYRTLLLGITKTSLNTQSF--ISEASFQETA 1271
YR L+ IT+ +N + SF+ET
Sbjct: 391 ------------------------------YRGHLMAITRHGINRNETGPMMRCSFEETV 480
Query: 1272 RVLAKAALRGRIDWLKGLKENLVLGGIIPVGTG 1304
+L AA+ D L+G+ EN++LG + P+GTG
Sbjct: 481 DILLDAAVYAETDHLRGVTENIMLGQLAPIGTG 579
>BQ752355 similar to PIR|A00694|RN DNA-directed RNA polymerase (EC 2.7.7.6)
III 160K chain - yeast (Saccharomyces cerevisiae),
partial (12%)
Length = 535
Score = 46.6 bits (109), Expect = 6e-05
Identities = 33/115 (28%), Positives = 59/115 (50%), Gaps = 20/115 (17%)
Frame = -2
Query: 130 IMSFSGARGNASQVHQLVGMRGLMSDPQGQMID------LP--------------IQSNL 169
+M+ SG++G+ V Q+V + G ++ D LP + ++
Sbjct: 453 VMAKSGSKGSEINVAQMVALVGQQIIGGKRVADGFQDRTLPHFYKNAPQPPSKGFVANSF 274
Query: 170 REGLSLTEYIISCYGARKGVVDTAVRTSDAGYLTRRLVEVVQHIVVRRTDCGTVR 224
GL TE+I R+G+VDTAV+T++ GY++RRL++ ++ + + D TVR
Sbjct: 273 YSGLLPTEFIFHAMSGREGLVDTAVKTAETGYMSRRLMKSLEDLSTQYDD--TVR 115
>TC82998 weakly similar to PIR|A00694|RNBY3L DNA-directed RNA polymerase (EC
2.7.7.6) III 160K chain - yeast (Saccharomyces
cerevisiae), partial (8%)
Length = 646
Score = 42.7 bits (99), Expect = 0.001
Identities = 34/143 (23%), Positives = 65/143 (44%)
Frame = +3
Query: 1169 LTIAQSRISLVNKIQRVYRSQGVHIHNRHIEIIVRQITSKVLVSEDGMSNVFSPGELIGL 1228
L I +R ++++IQ + G+ I RHI ++ +T + G+++G+
Sbjct: 75 LGIEAARYGIIDQIQYTMKQHGMSIDVRHIMLVADMMTVR--------------GQVLGM 212
Query: 1229 LRAQRTGRALEESICYRTLLLGITKTSLNTQSFISEASFQETARVLAKAALRGRIDWLKG 1288
R GI K +S + ASF+ + L A+LRG+ D ++G
Sbjct: 213 TRH------------------GIQKMG---RSALMLASFESSTDYLFDASLRGKGDPIEG 329
Query: 1289 LKENLVLGGIIPVGTGFKKIGDR 1311
+ + +++G I +GTG ++ R
Sbjct: 330 VSDCIIMGKPIHIGTGMIEVKQR 398
>TC91162 similar to PIR|T06754|T06754 DNA-directed RNA polymerase I 190K
chain homolog F15B8.150 - Arabidopsis thaliana, partial
(11%)
Length = 778
Score = 41.6 bits (96), Expect = 0.002
Identities = 33/110 (30%), Positives = 51/110 (46%), Gaps = 20/110 (18%)
Frame = +3
Query: 126 NPVHIMSFSGARG---NASQVHQLVGMRGLMSDPQGQMID---LP--------------I 165
N + +M+ SGA+G N Q+ +G + L +M+ LP I
Sbjct: 246 NWISLMTTSGAKGSMVNFQQISSHLGQQELEGKRVPRMVSGKTLPCFPSWDCSPRAGGFI 425
Query: 166 QSNLREGLSLTEYIISCYGARKGVVDTAVRTSDAGYLTRRLVEVVQHIVV 215
L EY C R+G+VDTAV+TS +GYL R L++ ++ + V
Sbjct: 426 IDRFLTALRPQEYYFHCMAGREGLVDTAVKTSRSGYLQRCLMKNLESLKV 575
>BQ751034 similar to GP|18307453|e probable DNA-directed RNA polymerase I
{Neurospora crassa}, partial (13%)
Length = 791
Score = 38.5 bits (88), Expect = 0.017
Identities = 31/116 (26%), Positives = 57/116 (48%), Gaps = 24/116 (20%)
Frame = +1
Query: 124 PFNPVHIMSFSGARG---NASQVHQLVGMRGLMSDPQGQMIDL-----------PIQSNL 169
P N + M+ SGA+G NA+ + +G + L +G+ + L P ++++
Sbjct: 190 PKNQMQSMTTSGAKGSSVNANLISCNLGQQVL----EGRRVPLMVSGKSLPCFKPFETDV 357
Query: 170 REG----------LSLTEYIISCYGARKGVVDTAVRTSDAGYLTRRLVEVVQHIVV 215
R G + EY R+G++DTAV+TS +GYL R +++ ++ + V
Sbjct: 358 RAGGYIVQRFLTGIRPQEYYFHHMAGREGLIDTAVKTSRSGYLQRCIIKGMEGLTV 525
>TC82897 similar to GP|16604396|gb|AAL24204.1 AT5g18630/T1A4_10 {Arabidopsis
thaliana}, partial (35%)
Length = 678
Score = 32.3 bits (72), Expect = 1.3
Identities = 16/43 (37%), Positives = 24/43 (55%)
Frame = +2
Query: 1090 ATIHGHYGEMLYEGDILVTFIYEKSRSGDITQGLPKVEQVLEV 1132
ATI Y +Y D+ F + SR GD+T+G +E V++V
Sbjct: 224 ATILVEYASAVYLSDLTQLFTWTCSRCGDLTKGFEIIELVVDV 352
>BE249090
Length = 647
Score = 30.4 bits (67), Expect = 4.8
Identities = 22/72 (30%), Positives = 38/72 (52%)
Frame = -3
Query: 633 IKVDRFFFIPQEVHILSESSSIMVRNNSIIGVNTPITLNKKSRVGGLVRVEKNKKKIELK 692
IKV+ F ++ ++ L + ++ + I+G+ LNK V G R++KN+K IE +
Sbjct: 213 IKVEVFIYL*EK---LVTTHKLIQVQH*ILGLTNIDELNK*EMVKGDNRIKKNRKVIERR 43
Query: 693 IFSGDIHFPGEI 704
I + GEI
Sbjct: 42 IIESFVT*KGEI 7
>TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (44%)
Length = 955
Score = 29.6 bits (65), Expect = 8.1
Identities = 20/57 (35%), Positives = 28/57 (49%), Gaps = 3/57 (5%)
Frame = -1
Query: 879 NPFFSIYKENIQQSLSQ---KHGTIRMLLNRNKENRSFIILSSSNCFQMGPFNNVKY 932
+PFF+ K NI+ S SQ KH +RSF+I + SNC NN ++
Sbjct: 760 DPFFNSKKRNIESSTSQVKNKHVLFAF-------SRSFLIQTISNCSSSRLINNPQH 611
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 41,836,444
Number of Sequences: 36976
Number of extensions: 624195
Number of successful extensions: 3431
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 3313
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3424
length of query: 1332
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1224
effective length of database: 5,021,319
effective search space: 6146094456
effective search space used: 6146094456
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Description of BAB33196.1

